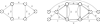A novel min-cost flow method for estimating transcript expression with RNA-Seq - PubMed (original) (raw)
A novel min-cost flow method for estimating transcript expression with RNA-Seq
Alexandru I Tomescu et al. BMC Bioinformatics. 2013.
Abstract
Background: Through transcription and alternative splicing, a gene can be transcribed into different RNA sequences (isoforms), depending on the individual, on the tissue the cell is in, or in response to some stimuli. Recent RNA-Seq technology allows for new high-throughput ways for isoform identification and quantification based on short reads, and various methods have been put forward for this non-trivial problem.
Results: In this paper we propose a novel radically different method based on minimum-cost network flows. This has a two-fold advantage: on the one hand, it translates the problem as an established one in the field of network flows, which can be solved in polynomial time, with different existing solvers; on the other hand, it is general enough to encompass many of the previous proposals under the least sum of squares model. Our method works as follows: in order to find the transcripts which best explain, under a given fitness model, a splicing graph resulting from an RNA-Seq experiment, we find a min-cost flow in an offset flow network, under an equivalent cost model. Under very weak assumptions on the fitness model, the optimal flow can be computed in polynomial time. Parsimoniously splitting the flow back into few path transcripts can be done with any of the heuristics and approximations available from the theory of network flows. In the present implementation, we choose the simple strategy of repeatedly removing the heaviest path.
Conclusions: We proposed a new very general method based on network flows for a multiassembly problem arising from isoform identification and quantification with RNA-Seq. Experimental results on prediction accuracy show that our method is very competitive with popular tools such as Cufflinks and IsoLasso. Our tool, called Traph (Transcrips in gRAPHs), is available at: http://www.cs.helsinki.fi/gsa/traph/.
Figures
Figure 1
Example of an offset network. An input G to Problem UTEJC (a), and the offset network G* (b); arcs are labeled with their capacity, unlabeled arcs having infinite capacity
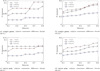
Figure 2
Performance on simulated data. Performance of IsoLasso, Cufflinks, and Traph on simulated data: single genes scenario (a), (b); batch mode scenario (c), (d)
Figure 3
Results on real human data. Histogram of the distribution of transcript lengths of the annotation, and of the ones reported by Traph, Cufflinks and IsoLasso
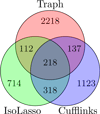
Figure 4
Results on real human data. Venn diagram of the intersections of the sets of reported transcripts
Similar articles
- ORMAN: optimal resolution of ambiguous RNA-Seq multimappings in the presence of novel isoforms.
Dao P, Numanagić I, Lin YY, Hach F, Karakoc E, Donmez N, Collins C, Eichler EE, Sahinalp SC. Dao P, et al. Bioinformatics. 2014 Mar 1;30(5):644-51. doi: 10.1093/bioinformatics/btt591. Epub 2013 Oct 15. Bioinformatics. 2014. PMID: 24130305 - Efficient RNA isoform identification and quantification from RNA-Seq data with network flows.
Bernard E, Jacob L, Mairal J, Vert JP. Bernard E, et al. Bioinformatics. 2014 Sep 1;30(17):2447-55. doi: 10.1093/bioinformatics/btu317. Epub 2014 May 9. Bioinformatics. 2014. PMID: 24813214 Free PMC article. - TIGAR: transcript isoform abundance estimation method with gapped alignment of RNA-Seq data by variational Bayesian inference.
Nariai N, Hirose O, Kojima K, Nagasaki M. Nariai N, et al. Bioinformatics. 2013 Sep 15;29(18):2292-9. doi: 10.1093/bioinformatics/btt381. Epub 2013 Jul 2. Bioinformatics. 2013. PMID: 23821651 - NURD: an implementation of a new method to estimate isoform expression from non-uniform RNA-seq data.
Ma X, Zhang X. Ma X, et al. BMC Bioinformatics. 2013 Jul 10;14:220. doi: 10.1186/1471-2105-14-220. BMC Bioinformatics. 2013. PMID: 23837734 Free PMC article. - FDM: a graph-based statistical method to detect differential transcription using RNA-seq data.
Singh D, Orellana CF, Hu Y, Jones CD, Liu Y, Chiang DY, Liu J, Prins JF. Singh D, et al. Bioinformatics. 2011 Oct 1;27(19):2633-40. doi: 10.1093/bioinformatics/btr458. Epub 2011 Aug 8. Bioinformatics. 2011. PMID: 21824971 Free PMC article.
Cited by
- Transcriptomic landscape of quiescent and proliferating human corneal stromal fibroblasts.
Kumar R, Tripathi R, Sinha NR, Mohan RR. Kumar R, et al. Exp Eye Res. 2024 Nov;248:110073. doi: 10.1016/j.exer.2024.110073. Epub 2024 Sep 5. Exp Eye Res. 2024. PMID: 39243928 - RNA-Seq Analysis Unraveling Novel Genes and Pathways Influencing Corneal Wound Healing.
Kumar R, Tripathi R, Sinha NR, Mohan RR. Kumar R, et al. Invest Ophthalmol Vis Sci. 2024 Sep 3;65(11):13. doi: 10.1167/iovs.65.11.13. Invest Ophthalmol Vis Sci. 2024. PMID: 39240550 Free PMC article. - Floria: fast and accurate strain haplotyping in metagenomes.
Shaw J, Gounot JS, Chen H, Nagarajan N, Yu YW. Shaw J, et al. Bioinformatics. 2024 Jun 28;40(Suppl 1):i30-i38. doi: 10.1093/bioinformatics/btae252. Bioinformatics. 2024. PMID: 38940183 Free PMC article. - A safety framework for flow decomposition problems via integer linear programming.
Dias FHC, Cáceres M, Williams L, Mumey B, Tomescu AI. Dias FHC, et al. Bioinformatics. 2023 Nov 1;39(11):btad640. doi: 10.1093/bioinformatics/btad640. Bioinformatics. 2023. PMID: 37862229 Free PMC article. - Phables: from fragmented assemblies to high-quality bacteriophage genomes.
Mallawaarachchi V, Roach MJ, Decewicz P, Papudeshi B, Giles SK, Grigson SR, Bouras G, Hesse RD, Inglis LK, Hutton ALK, Dinsdale EA, Edwards RA. Mallawaarachchi V, et al. Bioinformatics. 2023 Oct 3;39(10):btad586. doi: 10.1093/bioinformatics/btad586. Bioinformatics. 2023. PMID: 37738590 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources