Mutational analysis of the Pseudomonas aeruginosa myovirus KZ morphogenetic protease gp175 - PubMed (original) (raw)
Mutational analysis of the Pseudomonas aeruginosa myovirus KZ morphogenetic protease gp175
Julie A Thomas et al. J Virol. 2013 Aug.
Abstract
Pseudomonas aeruginosa myovirus KZ has a 270-kb genome within a T=27 icosahedral capsid that contains a large, unusual, and structurally well-defined protein cylindrical inner body (IB) spanning its interior. Proteolysis forms a pivotal stage in KZ head and IB morphogenesis, with the protease gp175 cleaving at least 19 of 49 different head proteins, including the major capsid protein and five major structural IB proteins. Here we show that the purified mature form of gp175 is active and cleaves purified IB structural proteins gp93 and gp89. Expression vector synthesis and purification of the zymogen/precursor yielded an active, mature-length protease, showing independent C-terminal gp175 self-cleavage autoactivation. Mutation of either the predicted catalytic serine or histidine inactivated mature gp175, supporting its classification as a serine protease and representing the first such direct biochemical demonstration with purified protease and substrate proteins for any phage protease. These mutations also blocked self-cleavage of the precursor while allowing intermolecular gp175 processing. To confirm the cleavage specificity of gp175, we mutated three cleavage sites in gp93, which blocked proteolysis at these sites. The N-terminal propeptide of gp93 was shown to undergo more extensive proteolysis than previously identified. We found that proteolysis in gp93 progressed from the N to C terminus, while blocking cleavage sites slowed but did not eliminate downstream proteolysis. These findings were shown by informatics to be relevant to the head morphogenesis of numbers of other related IB-containing giant phages as well as to T4 and herpesviruses, which have homologous proteases.
Figures
Fig 1
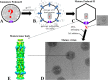
Schematic of morphogenesis of the ϕKZ head. (A) Immature prohead I, composed of 49 different proteins. (B) The protease gp175 undergoes autoactivation and cleaves 19 head proteins. (C) Processed prohead II contains all the proteins found in the mature capsid, including six abundant IB proteins. A structure which may represent an immature form of the IB, or breakdown product of it, has been observed by cryo-electron microscopy (cryo-EM) of prohead II. (D) Cryo-EM image of mature ϕKZ particles, showing IB bubbles in the head. (E) Reconstruction of the mature IB (previously published in reference and provided here with permission of the publisher).
Fig 2
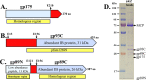
(A to C) Representation of ϕKZ gene products used in this study. Blue indicates regions present in the mature head. Red indicates precursor polypeptide regions that are processed and removed from the head. Protease cleavage sites previously identified by mass spectrometry are indicated with arrows (14). Yellow boxes indicate the most highly conserved region(s) in homologous proteins in related phages. (A) gp175, the prohead protease, and major IB proteins gp93 (B), and gp89 (C). Both gp93 and 89 are estimated to be present at about 100 copies per particle (14). (D) SDS-PAGE of purified ϕKZ heads obtained from a tailless mutant. The location of the forms of gp89, gp93, and gp175 in the mature head (gp89N, gp89C, gp93C, and gp175) as determined by mass spectral analysis (14). The N-terminal fragment of gp89 is not marked, as it migrates below the scope of this gel. MCP refers to the major capsid protein, present at 1,560 copies per virion.
Fig 3
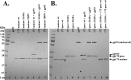
SDS-PAGE of activity assay products of expressed and purified mature and precursor forms of the ϕKZ protease, gp175, on IB protein gp93. (A) Assay of wild-type and mutated forms of mature length gp175. Lanes: 1, gp93; 2, gp175 expressed from a gene encoding the mature length of the wild-type protein (residues 1 to 210); 3, mature-length gp175 with mutated catalytic serine; 4, mature-length gp175 with a mutated catalytic histidine; lanes 5 to 7, products of assays of the wild-type and catalytic mutant forms of gp175 with gp93. (B) Assay products of wild-type and mutant forms of precursor-length gp175. Lanes: 1, purified gp175 expressed from a gene encoding the mature length of the wild-type protein (residues 1 to 210); 2, gp175 expressed from a gene encoding the zymogen or precursor form of the protease (residues 1 to 270); the bracket indicates fragments potentially representing inactivated forms of the protease (see Discussion); 3, gp175 expressed from a gene encoding the precursor form of gp175 with mutated catalytic serine; 4, gp175 expressed from a gene encoding the precursor form of gp175 with mutated catalytic histidine; 5, gp93. Lanes 6 to 8 show assay products of wild-type and catalytic residue mutants of gp175 expressed from the precursor-length gene with gp93. Lanes: 6, wild-type gp175; 7, catalytic serine mutant gp175; 8, catalytic histidine mutant gp175. The black circle indicates the C-terminal fragment of gp93 resulting from cleavage at E-156. The star indicates an N-terminal fragment of gp93. Lanes 9 and 10: assay products of wild-type gp175 expressed from a gene encoding the precursor form of gp175 with inactivated, precursor forms of gp175 (lane 9, catalytic serine mutant; lane 10, catalytic histidine mutant). wt, wild type prec., precursor.
Fig 4
SDS-PAGE of assay products of the ϕKZ protease gp175 on different substrates and under different assay conditions. (A) Lanes: 1, gp93; 2, gp93 and gp175; 3 to 6, gp175 and gp93. The assay was for 30 min at 37°C. Lane 3, gp175 preincubated for 30 min at 37°C immediately prior to the assay; lane 4, gp175 boiled for 2 min immediately, then quickly cooled on ice prior to the assay; lanes 5 and 6, products of assay mixtures supplemented with 0.1 and 1.0 mM serine protease inhibitor PMSF, respectively. (B) Lane 1, gp93 that had been boiled for 3 min and then quickly cooled on ice immediately prior to the assay, as a negative control with no protease added. Lane 2, gp93 that had been boiled prior to the assay with gp175 (not boiled). (C) Assay of gp175 on BSA. Lanes: 1, gp175; 2, BSA; 3, gp175 and BSA; 4, gp93; 5, gp175 and gp93.
Fig 5
Proteolysis of the ϕKZ IB protein gp89 by the protease gp175. (A) SDS-PAGE of assay products of gp89 with increasing concentrations of gp175 (lanes 1 to 5). Mature-length N- and C-terminal fragments of gp89 are indicated as black circles. Immature-length N- and C-terminal fragments of gp89 are indicated as blue circles, and the putative sites cleaved to produce them are indicated in blue. (B) Schematic of gp89, showing cleavage sites in addition to those identified by mass spectrometry (E-121 and E-156).
Fig 6
SDS-PAGE of assay products of ϕKZ protease gp175 on gp93. Lanes: 1, gp93; 2 to 10, increasing amounts of gp175 (a 2-fold dilution series from ∼2 μg to 2 ng in reverse) from assays (37°C for 30 min) with gp93 (∼2 μg); 11, gp175. Bands expected to represent C-terminal fragments of gp93 resulting from cleavage at E-13, E-137, and E-156 are indicated with a black square, black triangle, and black circle, respectively. Stars indicate fragments produced from proteolysis of the N-terminal propeptide of gp93. The bracket indicates fragments potentially representing inactivated forms of the protease (see Discussion).
Fig 7
Mutational analyses of ϕKZ gp93 protease cleavage sites. (A) Map of gp93, indicating propeptide versus mature regions and cleavage sites, confirmed by mutational analyses. Five sequences that conform to the cleavage motif are marked on the underside of the region and represent the propeptide region of gp93, and 10 other glutamate residues that do not contain the cleavage motif are also indicated. The sequence ALE-171 is marked with a cross to indicate that it was not cleaved in these studies. (B) SDS-PAGE of assay products of the ϕKZ protease with constructs of gp93 that have single, or combinations of, mutated cleavage sites (the sites identified by mass spectrometry, E13 and E156, and a predicted cleavage site, E137). Mutated residues are marked at the top of each lane. Bands expected to represent C-terminal fragments of gp93 resulting from cleavage at E-13, E-137, and E-156 are indicated with a black square, black triangle, and black circle, respectively. A band expected to represent a C-terminal fragment of gp93, for which the cleavage site was not determined, is indicated with a white circle. Cleavage fragments originating from the N-terminal propeptide region are indicated by black stars.
Fig 8
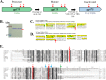
Autoinactivation of ϕKZ gp175. (A) Schematic of different forms of ϕKZ gp175 caused by the known maturation autocleavage and predicted inactivating autocleavage(s). Catalytic triad residues are indicated by red stars. (B) Enlarged section of lanes 1 and 2 from Fig. 3B, showing minor bands (red bracket) beneath the major band representing the mature form of gp175. (C) Mass spectral peptide coverage (yellow) of ϕKZ gp175 from a sample of cesium chloride gradient-purified heads, showing the peptide coverage ending at R-179 (red arrow) upstream of the maturation site (E-210). (D) Mass spectral peptide coverage of ϕKZ gp175 from a sample of ϕKZ heads, showing the peptide coverage ending at the maturation cleavage site, E-210 (red arrow). This sample was only subjected to differential centrifugation and is expected to contain more precursor particles and assembly intermediates than purified heads. (E) Alignment of the ϕKZ protease, gp175, with homologs in Pseudomonas phages 201ϕ2-1 (gp268), ϕPA3 (gp205), OBP (gp283), and EL (gp192) and homologs in Salmonella phage SPN3US (gp245), Erwinia phage PhiEaH2 (gp165), and Halocynthia phage JM-2012 (gp80). Homologs in uncultured bacteria are indicated by ub; the GenBank identifier for ub1 is EKD22713, and the GenBank identifier for ub2 is EKD89709. Catalytic histidine and serine residues are indicated in red and marked with stars, as is the candidate for the third catalytic residue (D-191). The ϕKZ gp175 maturation cleavage site (E-210) is indicated with a green arrow. Putative cleavage sites for ϕKZ gp175 self-inactivation (E-186 and E-195) are indicated with blue arrows.
Similar articles
- Extensive proteolysis of head and inner body proteins by a morphogenetic protease in the giant Pseudomonas aeruginosa phage φKZ.
Thomas JA, Weintraub ST, Wu W, Winkler DC, Cheng N, Steven AC, Black LW. Thomas JA, et al. Mol Microbiol. 2012 Apr;84(2):324-39. doi: 10.1111/j.1365-2958.2012.08025.x. Epub 2012 Mar 20. Mol Microbiol. 2012. PMID: 22429790 Free PMC article. - Genomic and proteomic analyses of the terminally redundant genome of the Pseudomonas aeruginosa phage PaP1: establishment of genus PaP1-like phages.
Lu S, Le S, Tan Y, Zhu J, Li M, Rao X, Zou L, Li S, Wang J, Jin X, Huang G, Zhang L, Zhao X, Hu F. Lu S, et al. PLoS One. 2013 May 13;8(5):e62933. doi: 10.1371/journal.pone.0062933. Print 2013. PLoS One. 2013. PMID: 23675441 Free PMC article. - Inhibition of Pseudomonas aeruginosa virulence: characterization of the AprA-AprI interface and species selectivity.
Bardoel BW, van Kessel KP, van Strijp JA, Milder FJ. Bardoel BW, et al. J Mol Biol. 2012 Jan 20;415(3):573-83. doi: 10.1016/j.jmb.2011.11.039. Epub 2011 Nov 29. J Mol Biol. 2012. PMID: 22154939 - [Phenogenetic characterization of a group of giant Phi KZ-like bacteriophages of Pseudomonas aeruginosa].
Burkal'tseva MV, Krylov VN, Pleteneva EA, Shaburova OV, Krylov SV, Volkart G, Sykilinda NN, Kurochkina LP, Mesianzhinov VV. Burkal'tseva MV, et al. Genetika. 2002 Nov;38(11):1470-9. Genetika. 2002. PMID: 12500672 Russian. - Probing the Activity of Eukaryotic Rhomboid Proteases In Vitro.
Cordier B, Lemberg MK. Cordier B, et al. Methods Enzymol. 2017;584:99-126. doi: 10.1016/bs.mie.2016.09.045. Epub 2016 Oct 27. Methods Enzymol. 2017. PMID: 28065274 Review.
Cited by
- Identification and characterization of phage protein and its activity against two strains of multidrug-resistant Pseudomonas aeruginosa.
Al-Wrafy F, Brzozowska E, Górska S, Drab M, Strus M, Gamian A. Al-Wrafy F, et al. Sci Rep. 2019 Sep 17;9(1):13487. doi: 10.1038/s41598-019-50030-5. Sci Rep. 2019. PMID: 31530875 Free PMC article. - Molecular architecture of tailed double-stranded DNA phages.
Fokine A, Rossmann MG. Fokine A, et al. Bacteriophage. 2014 Jan 1;4(1):e28281. doi: 10.4161/bact.28281. Epub 2014 Feb 21. Bacteriophage. 2014. PMID: 24616838 Free PMC article. Review. - A Cut above the Rest: Characterization of the Assembly of a Large Viral Icosahedral Capsid.
Reilly ER, Abajorga MK, Kiser C, Mohd Redzuan NH, Haidar Z, Adams LE, Diaz R, Pinzon JA, Hudson AO, Black LW, Hsia RC, Weintraub ST, Thomas JA. Reilly ER, et al. Viruses. 2020 Jul 5;12(7):725. doi: 10.3390/v12070725. Viruses. 2020. PMID: 32635654 Free PMC article. - Common Evolutionary Origin of Procapsid Proteases, Phage Tail Tubes, and Tubes of Bacterial Type VI Secretion Systems.
Fokine A, Rossmann MG. Fokine A, et al. Structure. 2016 Nov 1;24(11):1928-1935. doi: 10.1016/j.str.2016.08.013. Epub 2016 Sep 22. Structure. 2016. PMID: 27667692 Free PMC article. - Mass Spectral Analyses of Salmonella Myovirus SPN3US Reveal Conserved and Divergent Themes in Proteolytic Maturation of Large Icosahedral Capsids.
Scheuch A, Moran SAM, Faraone JN, Unwin SR, Vu G, Benítez AD, Mohd Redzuan NH, Molleur D, Pardo S, Weintraub ST, Thomas JA. Scheuch A, et al. Viruses. 2023 Mar 10;15(3):723. doi: 10.3390/v15030723. Viruses. 2023. PMID: 36992431 Free PMC article.
References
- Krylov VN, Dela Cruz DM, Hertveldt K, Ackermann HW. 2007. “ϕKZ-like viruses,” a proposed new genus of myovirus bacteriophages. Arch. Virol. 152:1955–1959 - PubMed
- Briers Y, Volckaert G, Cornelissen A, Lagaert S, Michiels CW, Hertveldt K, Lavigne R. 2007. Muralytic activity and modular structure of the endolysins of Pseudomonas aeruginosa bacteriophages ϕKZ and EL. Mol. Microbiol. 65:1334–1344 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources