Scalable web services for the PSIPRED Protein Analysis Workbench - PubMed (original) (raw)
. 2013 Jul;41(Web Server issue):W349-57.
doi: 10.1093/nar/gkt381. Epub 2013 Jun 8.
Affiliations
- PMID: 23748958
- PMCID: PMC3692098
- DOI: 10.1093/nar/gkt381
Scalable web services for the PSIPRED Protein Analysis Workbench
Daniel W A Buchan et al. Nucleic Acids Res. 2013 Jul.
Abstract
Here, we present the new UCL Bioinformatics Group's PSIPRED Protein Analysis Workbench. The Workbench unites all of our previously available analysis methods into a single web-based framework. The new web portal provides a greatly streamlined user interface with a number of new features to allow users to better explore their results. We offer a number of additional services to enable computationally scalable execution of our prediction methods; these include SOAP and XML-RPC web server access and new HADOOP packages. All software and services are available via the UCL Bioinformatics Group website at http://bioinf.cs.ucl.ac.uk/.
Figures
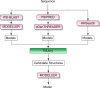
Figure 1.
Flowchart of the BioSerf2 automated homology modelling protocol. Incoming query sequences are independently matched to PDB chains using PSIBLAST, pGenTHREADER and HH. The three sets of models produced are then compared by the TMJury process, which produces up to 10 candidate homologous structures. These structures and their alignments to the query sequence are used as input for MODELLER to produce a single final structure.
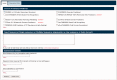
Figure 2.
The new front page of the PSIPRED Protein Analysis Workbench. The ‘Choose Prediction Method’ allows users to select any set of available analysis methods. A series of tabs appear along the top bar when users select analysis methods. These additional tabs allow users to select more detailed options.
Figure 3.
A typical results page where a user has selected all the available analysis methods. The image shows the analysis summary front page of the results. (a) Tab bar: this region contains a range of tabs the users can select to explore the detailed results from each analysis method. (b) Secondary structure map: this area of the page lays out the query sequence and colours residues as per the annotations made by each analysis methods. If users have selected a MEMSAT-SVM prediction, they can use the buttons provided to toggle between the different sets of sequence annotations. Here, α-helical residues are in pink, β-strand residues are in yellow and putative domain boundaries are indicated in blue. (c) Sequence resubmission widget: this region contains a cartoon selector that represents the query sequence. Users can use the sliders to select any sub-sequence of their query sequence and then select further analyses to perform on just the selected sub-sequence. (d) GenTHREADER summary: if a GenTHREADER analysis was calculated, the final region presents several schematic cartoons of each GenTHREADER alignment. Hits are presented as bars coloured as per the GenTHEADER confidence scores, green for greatest confidence, orange of moderate confidence and red for lowest confidence. If the user ‘mouses over’ the bars, a pop-up presents more detailed information about the alignment region.
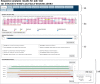
Figure 4.
The results summary for UniProt sequence A9DA50. (a) The secondary structure map showing the PSIPRED secondary structure predictions. α-Helical residues are in pink, β-strand residues are in yellow and putative domain boundaries are indicated in blue. (b) The pGenTHREADER alignments summary. Alignments in green have high-statistical confidence.
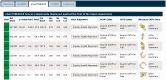
Figure 5.
The detailed pGenTHREADER results table for UniProt sequence A9DA50. The matched structures can all be seen to be similar all β-sheet structures. These are listed in CATH and PDBSum as either leucine-rich repeat domains or Tol-like receptors. Those chains fully classified in CATH list this domain in the 3.80.10-fold family.
Figure 6.
The MEMSAT-SVM results for the N-terminal region of UniProt sequence A9DA50. MEMSAT-SVM predicts a single membrane spanning helix (in purple) in this region.
Figure 7.
The results summary for the C-terminal region of UniProt sequence A9DA50. The secondary structure and pDomTHREADER results indicate that this region likely has a leading IG-like β-sheet domain.
Figure 8.
The FFPRED output for UniProt sequence A9DA50 indicating the best matched GO terms. Only the first 10 terms from the list are shown.
Similar articles
- Deep learning for the PSIPRED Protein Analysis Workbench.
Buchan DWA, Moffat L, Lau A, Kandathil SM, Jones DT. Buchan DWA, et al. Nucleic Acids Res. 2024 Jul 5;52(W1):W287-W293. doi: 10.1093/nar/gkae328. Nucleic Acids Res. 2024. PMID: 38747351 Free PMC article. - Protein structure prediction servers at University College London.
Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Bryson K, et al. Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W36-8. doi: 10.1093/nar/gki410. Nucleic Acids Res. 2005. PMID: 15980489 Free PMC article. - Protein annotation and modelling servers at University College London.
Buchan DW, Ward SM, Lobley AE, Nugent TC, Bryson K, Jones DT. Buchan DW, et al. Nucleic Acids Res. 2010 Jul;38(Web Server issue):W563-8. doi: 10.1093/nar/gkq427. Epub 2010 May 27. Nucleic Acids Res. 2010. PMID: 20507913 Free PMC article. - The PSIPRED Protein Analysis Workbench: 20 years on.
Buchan DWA, Jones DT. Buchan DWA, et al. Nucleic Acids Res. 2019 Jul 2;47(W1):W402-W407. doi: 10.1093/nar/gkz297. Nucleic Acids Res. 2019. PMID: 31251384 Free PMC article. - TMpro web server and web service: transmembrane helix prediction through amino acid property analysis.
Ganapathiraju M, Jursa CJ, Karimi HA, Klein-Seetharaman J. Ganapathiraju M, et al. Bioinformatics. 2007 Oct 15;23(20):2795-6. doi: 10.1093/bioinformatics/btm398. Epub 2007 Aug 27. Bioinformatics. 2007. PMID: 17724062 Free PMC article.
Cited by
- RTA1 Is Involved in Resistance to 7-Aminocholesterol and Secretion of Fungal Proteins in Cryptococcus neoformans.
Smith-Peavler ES, Patel R, Onumajuru AM, Bowring BG, Miller JL, Brunel JM, Djordjevic JT, Prabu MM, McClelland EE. Smith-Peavler ES, et al. Pathogens. 2022 Oct 26;11(11):1239. doi: 10.3390/pathogens11111239. Pathogens. 2022. PMID: 36364991 Free PMC article. - A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
Baßler J, Paternoga H, Holdermann I, Thoms M, Granneman S, Barrio-Garcia C, Nyarko A, Lee W, Stier G, Clark SA, Schraivogel D, Kallas M, Beckmann R, Tollervey D, Barbar E, Sinning I, Hurt E. Baßler J, et al. J Cell Biol. 2015 Jul 6;210(1):169-70. doi: 10.1083/jcb.20140811106112015c. J Cell Biol. 2015. PMID: 26150393 Free PMC article. No abstract available. - Unfoldome variation upon plant-pathogen interactions: strawberry infection by Colletotrichum acutatum.
Baraldi E, Coller E, Zoli L, Cestaro A, Tosatto SC, Zambelli B. Baraldi E, et al. Plant Mol Biol. 2015 Sep;89(1-2):49-65. doi: 10.1007/s11103-015-0353-7. Epub 2015 Aug 6. Plant Mol Biol. 2015. PMID: 26245354 - The Drosophila Pericentrin-like-protein (PLP) cooperates with Cnn to maintain the integrity of the outer PCM.
Richens JH, Barros TP, Lucas EP, Peel N, Pinto DM, Wainman A, Raff JW. Richens JH, et al. Biol Open. 2015 Jul 8;4(8):1052-61. doi: 10.1242/bio.012914. Biol Open. 2015. PMID: 26157019 Free PMC article. - Outer Membrane Proteins Derived from Non-cyanobacterial Lineage Cover the Peptidoglycan of Cyanophora paradoxa Cyanelles and Serve as a Cyanelle Diffusion Channel.
Kojima S, Muramoto K, Kusano T. Kojima S, et al. J Biol Chem. 2016 Sep 16;291(38):20198-209. doi: 10.1074/jbc.M116.746131. Epub 2016 Aug 8. J Biol Chem. 2016. PMID: 27502278 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources