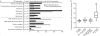Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation - PubMed (original) (raw)
Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation
Danielle L Swaney et al. Nat Methods. 2013 Jul.
Abstract
Cross-talk between different types of post-translational modifications on the same protein molecule adds specificity and combinatorial logic to signal processing, but it has not been characterized on a large-scale basis. We developed two methods to identify protein isoforms that are both phosphorylated and ubiquitylated in the yeast Saccharomyces cerevisiae, identifying 466 proteins with 2,100 phosphorylation sites co-occurring with 2,189 ubiquitylation sites. We applied these methods quantitatively to identify phosphorylation sites that regulate protein degradation via the ubiquitin-proteasome system. Our results demonstrate that distinct phosphorylation sites are often used in conjunction with ubiquitylation and that these sites are more highly conserved than the entire set of phosphorylation sites. Finally, we investigated how the phosphorylation machinery can be regulated by ubiquitylation. We found evidence for novel regulatory mechanisms of kinases and 14-3-3 scaffold proteins via proteasome-independent ubiquitylation.
Figures
Figure 1. Overview of methodology and modifications identified
Two sets of experiments were performed. The first was a qualitative assessment (non-SILAC), and the second, as illustrated here, was a SILAC experiment, in which cells from control and proteasome-inhibited cultures were mixed. We utilized two enrichment strategies for both experiments (a-b). (a) First, samples were enriched for Ub proteins. Non-Ub proteins (dashed lines) were digested and enriched for phosphopeptides. Ub proteins (solid lines) were further enriched for diGly-peptides and for phosphopeptides. (b) Proteins were digested to peptides and enriched for doubly charged peptides via strong-cation exchange (SCX). SCX fractions were then enriched for diGly-peptides prior to mass spectrometry analysis. (c-f) Illustration of four cases of how the same protein can exist in different combinations of modification states. (c) displays a phosphorylation site unique to the ubiquitylated protein isoform, while (d) and (e) display that some phosphorylation sites are found in both ubiquitylated and non-ubiquitylated isoforms. Finally, (f) displays a phosphorylation site unique to non-ubiquitylated proteins. (g) Overlap of phosphorylation sites and phosphoproteins from the Non-Ub (dashed lines) and Ub samples (solid lines).
Figure 2. Evolution and functional enrichment of modification sites
(a) GO enrichment analysis of ubiquitylated phosphoproteins vs. non-ubiquitylated phosphoproteins. GO enrichment was performed using Babelomics4. (b) For different populations of proteins we calculated the ratio of conserved phosphorylation sites over conserved amino acids from random sampling of the same number of phosphor-acceptor residues (Ser, Thr). Whiskers represent the lowest/highest data point within 1.5 times the interquartile range (*P = 0.0027, Kolmogorov-Smirnov test).
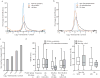
Figure 3. The effect of proteasome inhibition on protein and PTM site abundance, and properties of regulated phosphorylation sites
(a) Log2 distributions of proteins and ubiquitylation sites abundance changes in response to proteasome inhibition (50 μm Bortezomib for 1 hour). (b) Log2 distributions of phosphorylation and ubiquitylation sites abundance changes. (c) The distribution of sites matching a phosphodegron motif at various intervals of abundance changes after proteasome inhibition. (d) Half-life distributions for all protein identifications are compared to those proteins containing sites increasing in abundance by >75% (*P = 6.3 × 10−3), whiskers represent the 10th and 90th percentile. Protein half-life values were obtained from the literature. (e) The pairwise spatial distance (i.e. ångströms between alpha carbons) between either phosphorylation sites co-occurring with lysine ubiquitylation sites (Ub), or between the position of these same ubiquitylation sites to phosphorylation sites identified on non-ubiquitylated isoforms (Non-Ub). Distances between all pairs (white boxes) are compared to the distances between pairs for which both sites increase in abundance upon proteasome inhibition (grey boxes). (P < 0.0043). Whiskers represent the lowest/highest data point within 1.5 times the interquartile range.
Figure 4. Validation of a new phosphodegron and ubiquitin mediated regulation of phosphorylation machinery
(a-b) Yeast cells were treated with galactose to induce Swi5 expression and incubated with either cycloheximide to measure protein degradation (n = 3) (a) or Bortezomib to measure protein accumulation (b). Aliquots were taken at the indicated times and protein expression was monitored by immunoblotting with anti-HA to detect Swi5, or anti-α-tubulin for a loading control. Degradation levels measured from immunoblotting were quantified in triplicate, while Bortezomib induced accumulation was quantified from a single replicate. (b) Enrichment of ubiquitylation sites in the structure of the kinase domain. A representative structure pertaining to PDB ID: 1QMZ is shown. A total of 72 proteins showed enrichment of Ub sites within the Gly-rich loop or in Region A adjacent to the activation loop. Graph in the right represents the numeric values of enrichment over random for different positions within the kinase domain. Relevant regions within the domain are indicated. (c) Enrichment of ubiquitylation sites within the 14-3-3 phosphorylation-binding domain (PDB ID: 3MHR, 19 proteins from either human, mouse, or the yeast S. cerevisiae). The embedded table represents ubiquitylation sites identified in the phospho-binding and dimerization regions of the two 14-3-3 domain containing proteins in S. cerevisiae and the percent increase in abundance upon proteasome inhibition measured in this study.
Comment in
- Eavesdropping on PTM cross-talk through serial enrichment.
Webb K, Bennett EJ. Webb K, et al. Nat Methods. 2013 Jul;10(7):620-1. doi: 10.1038/nmeth.2526. Nat Methods. 2013. PMID: 23807195 No abstract available.
Similar articles
- Eavesdropping on PTM cross-talk through serial enrichment.
Webb K, Bennett EJ. Webb K, et al. Nat Methods. 2013 Jul;10(7):620-1. doi: 10.1038/nmeth.2526. Nat Methods. 2013. PMID: 23807195 No abstract available. - Studying Protein Ubiquitylation in Yeast.
Hovsepian J, Becuwe M, Kleifeld O, Glickman MH, Léon S. Hovsepian J, et al. Methods Mol Biol. 2016;1449:117-42. doi: 10.1007/978-1-4939-3756-1_5. Methods Mol Biol. 2016. PMID: 27613031 - Advances in characterizing ubiquitylation sites by mass spectrometry.
Sylvestersen KB, Young C, Nielsen ML. Sylvestersen KB, et al. Curr Opin Chem Biol. 2013 Feb;17(1):49-58. doi: 10.1016/j.cbpa.2012.12.009. Epub 2013 Jan 5. Curr Opin Chem Biol. 2013. PMID: 23298953 Review. - System-wide analysis reveals intrinsically disordered proteins are prone to ubiquitylation after misfolding stress.
Ng AH, Fang NN, Comyn SA, Gsponer J, Mayor T. Ng AH, et al. Mol Cell Proteomics. 2013 Sep;12(9):2456-67. doi: 10.1074/mcp.M112.023416. Epub 2013 May 28. Mol Cell Proteomics. 2013. PMID: 23716602 Free PMC article. - Protein ubiquitylation in pancreatic cancer.
Bonacci T, Roignot J, Soubeyran P. Bonacci T, et al. ScientificWorldJournal. 2010 Jul 20;10:1462-72. doi: 10.1100/tsw.2010.133. ScientificWorldJournal. 2010. PMID: 20661538 Free PMC article. Review.
Cited by
- Eavesdropping on PTM cross-talk through serial enrichment.
Webb K, Bennett EJ. Webb K, et al. Nat Methods. 2013 Jul;10(7):620-1. doi: 10.1038/nmeth.2526. Nat Methods. 2013. PMID: 23807195 No abstract available. - yRACK1/Asc1 proxiOMICs-Towards Illuminating Ships Passing in the Night.
Schmitt K, Valerius O. Schmitt K, et al. Cells. 2019 Nov 4;8(11):1384. doi: 10.3390/cells8111384. Cells. 2019. PMID: 31689955 Free PMC article. - Rab5 GTPases are required for optimal TORC2 function.
Locke MN, Thorner J. Locke MN, et al. J Cell Biol. 2019 Mar 4;218(3):961-976. doi: 10.1083/jcb.201807154. Epub 2018 Dec 21. J Cell Biol. 2019. PMID: 30578283 Free PMC article. - Recent findings and technological advances in phosphoproteomics for cells and tissues.
von Stechow L, Francavilla C, Olsen JV. von Stechow L, et al. Expert Rev Proteomics. 2015;12(5):469-87. doi: 10.1586/14789450.2015.1078730. Expert Rev Proteomics. 2015. PMID: 26400465 Free PMC article. Review. - Prediction of Functionally Important Phospho-Regulatory Events in Xenopus laevis Oocytes.
Johnson JR, Santos SD, Johnson T, Pieper U, Strumillo M, Wagih O, Sali A, Krogan NJ, Beltrao P. Johnson JR, et al. PLoS Comput Biol. 2015 Aug 27;11(8):e1004362. doi: 10.1371/journal.pcbi.1004362. eCollection 2015 Aug. PLoS Comput Biol. 2015. PMID: 26312481 Free PMC article.
References
- Jenuwein T, Allis CD. Translating the histone code. Science Signaling. 2001;293:1074. - PubMed
- Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Molecular Cell. 2007;28:730–738. - PubMed
- Ichimura T. 14-3-3 proteins modulate the expression of epithelial Na+ channels by phosphorylation-dependent interaction with Nedd4-2 ubiquitin ligase. J Biol Chem. 2004;280:13187–13194. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 GM103533/GM/NIGMS NIH HHS/United States
- R00 CA140789/CA/NCI NIH HHS/United States
- P50 GM082250/GM/NIGMS NIH HHS/United States
- R00CA140789/CA/NCI NIH HHS/United States
- P01 AI091575/AI/NIAID NIH HHS/United States
- P01 AI090935/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P50 GM081879/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases