Chemical modification-assisted bisulfite sequencing (CAB-Seq) for 5-carboxylcytosine detection in DNA - PubMed (original) (raw)
. 2013 Jun 26;135(25):9315-7.
doi: 10.1021/ja4044856. Epub 2013 Jun 17.
Affiliations
- PMID: 23758547
- PMCID: PMC3727251
- DOI: 10.1021/ja4044856
Chemical modification-assisted bisulfite sequencing (CAB-Seq) for 5-carboxylcytosine detection in DNA
Xingyu Lu et al. J Am Chem Soc. 2013.
Abstract
5-Methylcytosine (5mC) in DNA can be oxidized stepwise to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) by the TET family proteins. Thymine DNA glycosylase can further remove 5fC and 5caC, connecting 5mC oxidation with active DNA demethylation. Here, we present a chemical modification-assisted bisulfite sequencing (CAB-Seq) that can detect 5caC with single-base resolution in DNA. We optimized 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC)-catalyzed amide bond formation between the carboxyl group of 5caC and a primary amine group. We found that the modified 5caC can survive the bisulfite treatment without deamination. Therefore, this chemical labeling coupled with bisulfite treatment provides a base-resolution detection and sequencing method for 5caC.
Figures
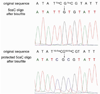
Figure 1
Sanger sequencing results of unlabeled 76 mer 5caC (upper panel) and EDC-labeled 5caC (bottom panel) after bisulfite treatment. The original sequence and the sequence after bisulfite were shown above the corresponding peak. In the bottom panel, the labeled DNA was not further purified before bisulfite treatment. The small background peak of T comes from unlabeled cytosine, which reflects the labeling efficiency.
Scheme 1. EDC-catalyzed chemical labeling of 5caC.a
_a_Designed labeling strategy for 5caC. The carboxyl group of 5caC in duplex DNA can be labeled by EDC-catalyzed coupling reaction to form biotin-S-S-5caC using biotin-S-S-amine.
Scheme 2. Proposed deamination mechanism of 5caC under bisulfite treatment
Similar articles
- Base-resolution profiling of active DNA demethylation using MAB-seq and caMAB-seq.
Wu H, Wu X, Zhang Y. Wu H, et al. Nat Protoc. 2016 Jun;11(6):1081-100. doi: 10.1038/nprot.2016.069. Epub 2016 May 12. Nat Protoc. 2016. PMID: 27172168 Free PMC article. - Methylation-assisted bisulfite sequencing to simultaneously map 5fC and 5caC on a genome-wide scale for DNA demethylation analysis.
Neri F, Incarnato D, Krepelova A, Parlato C, Oliviero S. Neri F, et al. Nat Protoc. 2016 Jul;11(7):1191-205. doi: 10.1038/nprot.2016.063. Epub 2016 Jun 9. Nat Protoc. 2016. PMID: 27281647 - Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing.
Wu H, Wu X, Shen L, Zhang Y. Wu H, et al. Nat Biotechnol. 2014 Dec;32(12):1231-40. doi: 10.1038/nbt.3073. Epub 2014 Nov 2. Nat Biotechnol. 2014. PMID: 25362244 Free PMC article. - Chemoselective labeling and site-specific mapping of 5-formylcytosine as a cellular nucleic acid modification.
Dietzsch J, Feineis D, Höbartner C. Dietzsch J, et al. FEBS Lett. 2018 Jun;592(12):2032-2047. doi: 10.1002/1873-3468.13058. Epub 2018 May 8. FEBS Lett. 2018. PMID: 29683490 Review. - Epigenetic modifications in DNA could mimic oxidative DNA damage: A double-edged sword.
Ito S, Kuraoka I. Ito S, et al. DNA Repair (Amst). 2015 Aug;32:52-57. doi: 10.1016/j.dnarep.2015.04.013. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25956859 Review.
Cited by
- Crossing epigenetic frontiers: the intersection of novel histone modifications and diseases.
Yao W, Hu X, Wang X. Yao W, et al. Signal Transduct Target Ther. 2024 Sep 16;9(1):232. doi: 10.1038/s41392-024-01918-w. Signal Transduct Target Ther. 2024. PMID: 39278916 Free PMC article. Review. - Epigenetic regulation in metabolic diseases: mechanisms and advances in clinical study.
Wu YL, Lin ZJ, Li CC, Lin X, Shan SK, Guo B, Zheng MH, Li F, Yuan LQ, Li ZH. Wu YL, et al. Signal Transduct Target Ther. 2023 Mar 2;8(1):98. doi: 10.1038/s41392-023-01333-7. Signal Transduct Target Ther. 2023. PMID: 36864020 Free PMC article. Review. - The Transition from Cancer "omics" to "epi-omics" through Next- and Third-Generation Sequencing.
Athanasopoulou K, Daneva GN, Boti MA, Dimitroulis G, Adamopoulos PG, Scorilas A. Athanasopoulou K, et al. Life (Basel). 2022 Dec 2;12(12):2010. doi: 10.3390/life12122010. Life (Basel). 2022. PMID: 36556377 Free PMC article. Review. - Labeling and sequencing nucleic acid modifications using bio-orthogonal tools.
Liu H, Wang Y, Zhou X. Liu H, et al. RSC Chem Biol. 2022 Jun 22;3(8):994-1007. doi: 10.1039/d2cb00087c. eCollection 2022 Aug 3. RSC Chem Biol. 2022. PMID: 35975003 Free PMC article. Review. - Protonation-Dependent Sequencing of 5-Formylcytidine in RNA.
Link CN, Thalalla Gamage S, Gallimore D, Kopajtich R, Evans C, Nance S, Fox SD, Andresson T, Chari R, Ivanic J, Prokisch H, Meier JL. Link CN, et al. Biochemistry. 2022 Apr 5;61(7):535-544. doi: 10.1021/acs.biochem.1c00761. Epub 2022 Mar 14. Biochemistry. 2022. PMID: 35285626 Free PMC article.
References
- Klose RJ, Bird AP. Trends Biochem. Sci. 2006;31:89. - PubMed
- Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, Carell T. Angew. Chem. Int. Ed. 2011;50:7008. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HG006827/HG/NHGRI NIH HHS/United States
- NS079625/NS/NINDS NIH HHS/United States
- R01 HG006827/HG/NHGRI NIH HHS/United States
- R01 NS079625/NS/NINDS NIH HHS/United States
- HD073162/HD/NICHD NIH HHS/United States
- R21 HD073162/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources