Genome-wide and species-wide in silico screening for intragenic MicroRNAs in human, mouse and chicken - PubMed (original) (raw)
Genome-wide and species-wide in silico screening for intragenic MicroRNAs in human, mouse and chicken
Irena Godnic et al. PLoS One. 2013.
Abstract
MicroRNAs (miRNAs) are non-coding RNAs (ncRNAs) involved in regulation of gene expression. Intragenic miRNAs, especially those exhibiting a high degree of evolutionary conservation, have been shown to be coordinately regulated and/or expressed with their host genes, either with synergistic or antagonistic correlation patterns. However, the degree of cross-species conservation of miRNA/host gene co-location is not known and co-expression information is incomplete and fragmented among several studies. Using the genomic resources (miRBase and Ensembl) we performed a genome-wide in silico screening (GWISS) for miRNA/host gene pairs in three well-annotated vertebrate species: human, mouse, and chicken. Approximately half of currently annotated miRNA genes resided within host genes: 53.0% (849/1,600) in human, 48.8% (418/855) in mouse, and 42.0% (210/499) in chicken, which we present in a central publicly available Catalog of intragenic miRNAs (http://www.integratomics-time.com/miR-host/catalog). The miRNA genes resided within either protein-coding or ncRNA genes, which include long intergenic ncRNAs (lincRNAs) and small nucleolar RNAs (snoRNAs). Twenty-seven miRNA genes were found to be located within the same host genes in all three species and the data integration from literature and databases showed that most (26/27) have been found to be co-expressed. Particularly interesting are miRNA genes located within genes encoding for miRNA silencing machinery (DGCR8, DICER1, and SND1 in human and Cnot3, Gdcr8, Eif4e, Tnrc6b, and Xpo5 in mouse). We furthermore discuss a potential for phenotype misattribution of miRNA host gene polymorphism or gene modification studies due to possible collateral effects on miRNAs hosted within them. In conclusion, the catalog of intragenic miRNAs and identified 27 miRNA/host gene pairs with cross-species conserved co-location, co-expression, and potential co-regulation, provide excellent candidates for further functional annotation of intragenic miRNAs in health and disease.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
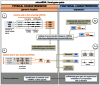
Figure 1. Workflow of the study.
GEA – Gene Expression Atlas.
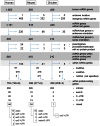
Figure 2. Diagram of genomic distribution of miRNA genes in human, mouse, and chicken.
* - microRNA genes overlapping protein-coding and ncRNA genes; mixed - microRNA genes overlapping intron, exon or UTR, depending on overlapping host gene transcripts. For details see online table:
http://www.integratomics-time.com/miR-host/catalog
.
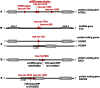
Figure 3. Examples of co-location of miRNA genes with protein-coding and ncRNA genes.
A) Protein-coding gene HTR2C with four resident miRNA genes, two of which form a cluster. B) A miRNA gene cluster located within lincRNA gene FTX. C) MicroRNA gene hsa-mir-10a located within two overlapping protein-coding genes. D) Overlapping miRNA gene (hsa-mir-664b) comprising a miR-seed-SNP, and snoRNA gene (SNORA36A) residing within protein-coding DKC1. E) Gene DGCR8, associated with miRNA biogenesis, hosts two miRNA genes, one of which comprises a miR-seed-SNP.
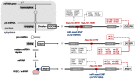
Figure 4. Cross talk of miRNA-related genomic elements.
Overlapping miRNA genes (hsa-mir-3618 and mir-1306, mir-3173, and mir-593), miRNA polymorphisms (miR-seed-SNPs (rs12159555 and rs73721294), host genes encoding for miRNA processing machinery components (DGCR8, DICER1, and SND1), miRNA target sites within host genes, and miRNAs targeting other host genes. Arrow with solid line: experimentally validated miRNA targets; arrow with dashed line: predicted miRNA targets.
Similar articles
- A potential role for intragenic miRNAs on their hosts' interactome.
Hinske LC, Galante PA, Kuo WP, Ohno-Machado L. Hinske LC, et al. BMC Genomics. 2010 Oct 1;11:533. doi: 10.1186/1471-2164-11-533. BMC Genomics. 2010. PMID: 20920310 Free PMC article. - miRIAD-integrating microRNA inter- and intragenic data.
Hinske LC, França GS, Torres HA, Ohara DT, Lopes-Ramos CM, Heyn J, Reis LF, Ohno-Machado L, Kreth S, Galante PA. Hinske LC, et al. Database (Oxford). 2014 Oct 6;2014:bau099. doi: 10.1093/database/bau099. Print 2014. Database (Oxford). 2014. PMID: 25288656 Free PMC article. - Gene silencing in vitro and in vivo using intronic microRNAs.
Lin SL, Ying SY. Lin SL, et al. Methods Mol Biol. 2006;342:295-312. doi: 10.1385/1-59745-123-1:295. Methods Mol Biol. 2006. PMID: 16957384 Review. - A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome.
Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E, Peterson KJ. Fromm B, et al. Annu Rev Genet. 2015;49:213-42. doi: 10.1146/annurev-genet-120213-092023. Epub 2015 Oct 14. Annu Rev Genet. 2015. PMID: 26473382 Free PMC article. Review.
Cited by
- Association study of a common genetic variant in pre-miR-1596 with chicken performance traits.
Li H, Tian Y, Sun G, Liu X, Jiang R, Han R, Li G, Kang X. Li H, et al. Mol Biol Rep. 2014 Nov;41(11):7175-81. doi: 10.1007/s11033-014-3600-0. Epub 2014 Jul 20. Mol Biol Rep. 2014. PMID: 25038725 - The Role of microRNAs in Epithelial Ovarian Cancer Metastasis.
Nguyen VHL, Yue C, Du KY, Salem M, O'Brien J, Peng C. Nguyen VHL, et al. Int J Mol Sci. 2020 Sep 25;21(19):7093. doi: 10.3390/ijms21197093. Int J Mol Sci. 2020. PMID: 32993038 Free PMC article. Review. - The role of hsa-miR-193a-5p as an important factor for control of inositol in alopecia areata.
AbdElneam AI, Al-Dhubaibi MS, Bahaj SS, Mohammed GF, Atef LM. AbdElneam AI, et al. Skin Res Technol. 2024 Jul;30(7):e13800. doi: 10.1111/srt.13800. Skin Res Technol. 2024. PMID: 38925555 Free PMC article. - Insights Into SND1 Oncogene Promoter Regulation.
Ochoa B, Chico Y, Martínez MJ. Ochoa B, et al. Front Oncol. 2018 Dec 11;8:606. doi: 10.3389/fonc.2018.00606. eCollection 2018. Front Oncol. 2018. PMID: 30619748 Free PMC article. Review. - Deciphering the Molecular Mechanism of Incurable Muscle Disease by a Novel Method for the Interpretation of miRNA Dysregulation.
Israeli D, Vu Hong A, Corre G, Miagoux Q, Richard I. Israeli D, et al. Noncoding RNA. 2022 Jun 30;8(4):48. doi: 10.3390/ncrna8040048. Noncoding RNA. 2022. PMID: 35893231 Free PMC article.
References
- Bartel DP (2004) MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 116: 281–297. - PubMed
- Lee Y, Ahn C, Han J, Choi H, Kim J, et al. (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous