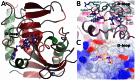Structural basis and selectivity of tankyrase inhibition by a Wnt signaling inhibitor WIKI4 - PubMed (original) (raw)
Structural basis and selectivity of tankyrase inhibition by a Wnt signaling inhibitor WIKI4
Teemu Haikarainen et al. PLoS One. 2013.
Abstract
Recently a novel inhibitor of Wnt signaling was discovered. The compound, WIKI4, was found to act through tankyrase inhibition and regulate β-catenin levels in many cancer cell lines and human embryonic stem cells. Here we confirm that WIKI4 is a high potency tankyrase inhibitor and that it selectively inhibits tankyrases over other ARTD enzymes tested. The binding mode of the compound to tankyrase 2 was determined by protein X-ray crystallography to 2.4 Å resolution. The structure revealed a novel binding mode to the adenosine subsite of the donor NAD(+) binding groove of the catalytic domain. Our results form a structural basis for further development of potent and selective tankyrase inhibitors based on the WIKI4 scaffold.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
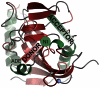
Figure 1. Structure of TNKS2 ARTD domain.
Acceptor and donor NAD+ binding sites, including nicotinamide subsite (NI) and adenosine subsite (ADE) are labelled.
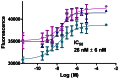
Figure 2. Potency of WIKI4 against TNKS1.
The in vitro dose response curves were measured three times with a fluorescence-based homogenous activity assay.
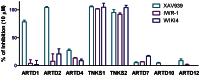
Figure 3. Profiling of inhibitor selectivity.
The selectivity of WIKI4 against 8 ARTDs polymerases was measured at 10 µM concentration. XAV939 and IWR-1 were used as controls.
Figure 4. Binding of WIKI4 to TNKS2.
a) An overview of TNKS2 structure showing the binding site of WIKI4 (lilac) and XAV939 (dark purple) (pdb accession code 3KR8). b) Comparison of apo TNKS2 structure (pink) (pdb accession code 3KR7) and WIKI4 (turquoise) bound structure of TNKS2. c) Surface electrostatic presentation of WIKI4 binding site. Positive (surface potential charge above 0.25 V) and negative (surface potential charge below −0.25 V) electrostatic regions are colored blue and red, respectively.
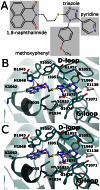
Figure 5. Interactions of WIKI4 with TNKS2 catalytic domain.
a) Chemical structure of WIKI4. b) Binding mode of WIKI4 to monomer A. c) Binding mode of WIKI4 to monomer B.
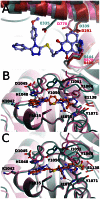
Figure 6. Comparison of WIKI4 binding to tankyrase 2 with other tankyrase selective inhibitors and with ARTD1-3 structures.
WIKI4 - TNKS2 protein structure is colored in turquoise and WIKI4 is colored in lilac. a) Comparison of the WIKI4 binding sites in TNKS2, ARTD1 (pink) (pdb accession code 3GJW), ARTD2 (green) (pdb accession code 3KCZ), and ARTD3 (red) (pdb accession code 3FHB). ARD, ARTD regulatory domain. b) Comparison of the binding of WIKI4 and IWR-1. Hydrogen bonds for WIKI4 and IWR-1 are shown in black and gray dotted lines, respectively. IWR-1 - TNKS2 protein structure is colored in pink and IWR-1 is colored in orange. c) Comparison of the binding of WIKI4 and G007-LK. Hydrogen bonds for WIKI4 and G007-LK are shown in black and gray dotted lines, respectively. G007-LK - TNKS2 protein structure is colored in pink and G007-LK is colored in orange.
Similar articles
- Novel binding mode of a potent and selective tankyrase inhibitor.
Gunaydin H, Gu Y, Huang X. Gunaydin H, et al. PLoS One. 2012;7(3):e33740. doi: 10.1371/journal.pone.0033740. Epub 2012 Mar 16. PLoS One. 2012. PMID: 22438990 Free PMC article. - WIKI4, a novel inhibitor of tankyrase and Wnt/ß-catenin signaling.
James RG, Davidson KC, Bosch KA, Biechele TL, Robin NC, Taylor RJ, Major MB, Camp ND, Fowler K, Martins TJ, Moon RT. James RG, et al. PLoS One. 2012;7(12):e50457. doi: 10.1371/journal.pone.0050457. Epub 2012 Dec 5. PLoS One. 2012. PMID: 23227175 Free PMC article. - Development and structural analysis of adenosine site binding tankyrase inhibitors.
Haikarainen T, Waaler J, Ignatev A, Nkizinkiko Y, Venkannagari H, Obaji E, Krauss S, Lehtiö L. Haikarainen T, et al. Bioorg Med Chem Lett. 2016 Jan 15;26(2):328-333. doi: 10.1016/j.bmcl.2015.12.018. Epub 2015 Dec 8. Bioorg Med Chem Lett. 2016. PMID: 26706174 - Tankyrases as drug targets.
Lehtiö L, Chi NW, Krauss S. Lehtiö L, et al. FEBS J. 2013 Aug;280(15):3576-93. doi: 10.1111/febs.12320. Epub 2013 Jun 18. FEBS J. 2013. PMID: 23648170 Review. - Zoning in on Tankyrases: A Brief Review on the Past, Present and Prospective Studies.
Peters XQ, Malinga TH, Agoni C, Olotu FA, Soliman MES. Peters XQ, et al. Anticancer Agents Med Chem. 2019;19(16):1920-1934. doi: 10.2174/1871520619666191019114321. Anticancer Agents Med Chem. 2019. PMID: 31648650 Review.
Cited by
- PARPs in genome stability and signal transduction: implications for cancer therapy.
Palazzo L, Ahel I. Palazzo L, et al. Biochem Soc Trans. 2018 Dec 17;46(6):1681-1695. doi: 10.1042/BST20180418. Epub 2018 Nov 12. Biochem Soc Trans. 2018. PMID: 30420415 Free PMC article. Review. - Pleiotropic roles of tankyrase/PARP proteins in the establishment and maintenance of human naïve pluripotency.
Zimmerlin L, Zambidis ET. Zimmerlin L, et al. Exp Cell Res. 2020 May 1;390(1):111935. doi: 10.1016/j.yexcr.2020.111935. Epub 2020 Mar 7. Exp Cell Res. 2020. PMID: 32151493 Free PMC article. Review. - Small-molecule inhibitors of Wnt signaling pathway: towards novel anticancer therapeutics.
Zheng S, Liu J, Wu Y, Huang TL, Wang G. Zheng S, et al. Future Med Chem. 2015;7(18):2485-505. doi: 10.4155/fmc.15.159. Epub 2015 Dec 16. Future Med Chem. 2015. PMID: 26670195 Free PMC article. Review. - Identifying Direct Protein Targets of Poly-ADP-Ribose Polymerases (PARPs) Using Engineered PARP Variants-Orthogonal Nicotinamide Adenine Dinucleotide (NAD+) Analog Pairs.
Carter-O'Connell I, Cohen MS. Carter-O'Connell I, et al. Curr Protoc Chem Biol. 2015 Jun 1;7(2):121-39. doi: 10.1002/9780470559277.ch140259. Curr Protoc Chem Biol. 2015. PMID: 26344237 Free PMC article. - The Poly(ADP-ribose) Polymerase Enzyme Tankyrase Antagonizes Activity of the β-Catenin Destruction Complex through ADP-ribosylation of Axin and APC2.
Croy HE, Fuller CN, Giannotti J, Robinson P, Foley AVA, Yamulla RJ, Cosgriff S, Greaves BD, von Kleeck RA, An HH, Powers CM, Tran JK, Tocker AM, Jacob KD, Davis BK, Roberts DM. Croy HE, et al. J Biol Chem. 2016 Jun 10;291(24):12747-12760. doi: 10.1074/jbc.M115.705442. Epub 2016 Apr 11. J Biol Chem. 2016. PMID: 27068743 Free PMC article.
References
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, et al. (2010) Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell 142: 943–953 doi:10.1016/j.cell.2010.08.016 - DOI - PubMed
- Guettler S, LaRose J, Petsalaki E, Gish G, Scotter A, et al. (2011) Structural basis and sequence rules for substrate recognition by Tankyrase explain the basis for cherubism disease. Cell 147: 1340–1354 doi:10.1016/j.cell.2011.10.046 - DOI - PubMed
- Smith S, Giriat I, Schmitt A, De Lange T (1998) Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282: 1484–1487 doi: 10.1126/science.282.5393.1484. - PubMed
- Sbodio JI, Lodish HF, Chi N-W (2002) Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase). Biochem J 361: 451–459 doi: 10.1042/0264-6021:3610451 - DOI - PMC - PubMed
- Riffell JL, Lord CJ, Ashworth A (2012) Tankyrase-targeted therapeutics: expanding opportunities in the PARP family. Nat Rev Drug Discov 11: 923–936 doi:10.1038/nrd3868 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
The work was funded by Biocenter Oulu (http://www.oulu.fi/biocenter/) and by Sigrid Jusélius foundation (http://www.sigridjuselius.fi/). MN and HV are members of the National Doctoral Programme of Informational and Structural Biology. The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007–2013) under BioStruct-X (grant agreement N°283570). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources