Oral microbiome of deep and shallow dental pockets in chronic periodontitis - PubMed (original) (raw)
Multicenter Study
Oral microbiome of deep and shallow dental pockets in chronic periodontitis
Xiuchun Ge et al. PLoS One. 2013.
Abstract
We examined the subgingival bacterial biodiversity in untreated chronic periodontitis patients by sequencing 16S rRNA genes. The primary purpose of the study was to compare the oral microbiome in deep (diseased) and shallow (healthy) sites. A secondary purpose was to evaluate the influences of smoking, race and dental caries on this relationship. A total of 88 subjects from two clinics were recruited. Paired subgingival plaque samples were taken from each subject, one from a probing site depth >5 mm (deep site) and the other from a probing site depth ≤3mm (shallow site). A universal primer set was designed to amplify the V4-V6 region for oral microbial 16S rRNA sequences. Differences in genera and species attributable to deep and shallow sites were determined by statistical analysis using a two-part model and false discovery rate. Fifty-one of 170 genera and 200 of 746 species were found significantly different in abundances between shallow and deep sites. Besides previously identified periodontal disease-associated bacterial species, additional species were found markedly changed in diseased sites. Cluster analysis revealed that the microbiome difference between deep and shallow sites was influenced by patient-level effects such as clinic location, race and smoking. The differences between clinic locations may be influenced by racial distribution, in that all of the African Americans subjects were seen at the same clinic. Our results suggested that there were influences from the microbiome for caries and periodontal disease and these influences are independent.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Shannon diversity index of genus- (A) and species-level (B) OTUs in the deep and shallow sites.
*, p-value<0.05; **, p-value<0.01.
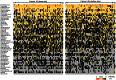
Figure 2. Genus-level OTUs with >1.0% of abundance in at least one patient in deep and/or shallow sites.
Colors reflect the abundance of OTUs from high (red) to low (black); gray, OTU missing.
Figure 3. Genus-level OTUs significantly different between shallow and deep pockets.
Of 170 genera, 51 showed a statistically significant difference in abundance (%) between shallow and deep sites. Data were statistically analyzed by the two-part model and false discovery rate (FDR). The FDR q-value of 51 genera was <0.05 which represents significant difference between shallow and deep samples. Inserted figure, phylum-level OTUs significantly different between shallow and deep sites; **, p-value <0.01; ***, p-value <0.001.
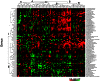
Figure 4. Clustering analysis of log2 ratio of deep abundance to the shallow at genus level using Hierarchical Trees (HCL).
The data were obtained using log2 ratio (deep abundance/shallow abundance). Both deep and shallow abundance had 0.00001 added due to the many zeros in the samples. The genera present in fewer than 10% of samples were excluded. The genera were clustered into two groups (I and II) and 88 patients were grouped into two types (A and B), each of which contains 2 subtypes (A1 and A2, B1 and B2).
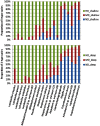
Figure 5. The species associated with dental caries.
The abundances of species-level OTUs were statistically analyzed in 3 levels of dental caries in deep and shallow sites, respectively. The species with a significant difference (α = 0.05) in 3 levels of dental caries were shown. HD_shallow, high decay and shallow site; MD_shallow, moderate decay and shallow site; ND_shallow, non-decay and shallow site; HD_deep, high decay and deep site; MD_ deep, moderate decay and deep site; ND_ deep, non-decay in deep site.
Similar articles
- The Subgingival Microbiome of Periodontal Pockets With Different Probing Depths in Chronic and Aggressive Periodontitis: A Pilot Study.
Shi M, Wei Y, Hu W, Nie Y, Wu X, Lu R. Shi M, et al. Front Cell Infect Microbiol. 2018 May 1;8:124. doi: 10.3389/fcimb.2018.00124. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29765908 Free PMC article. - Subgingival microbiota of Sri Lankan tea labourers naïve to oral hygiene measures.
Zhuang LF, Watt RM, Steiner S, Lang-Hua BH, Wang R, Ramseier CA, Lang NP. Zhuang LF, et al. J Clin Periodontol. 2014 May;41(5):433-41. doi: 10.1111/jcpe.12230. Epub 2014 Feb 26. J Clin Periodontol. 2014. PMID: 24460707 - Microbial analysis of subgingival plaque samples compared to that of whole saliva in patients with periodontitis.
Haririan H, Andrukhov O, Bertl K, Lettner S, Kierstein S, Moritz A, Rausch-Fan X. Haririan H, et al. J Periodontol. 2014 Jun;85(6):819-28. doi: 10.1902/jop.2013.130306. Epub 2013 Oct 21. J Periodontol. 2014. PMID: 24144271 - Investigation and quantification of key periodontal pathogens in patients with type 2 diabetes.
Field CA, Gidley MD, Preshaw PM, Jakubovics N. Field CA, et al. J Periodontal Res. 2012 Aug;47(4):470-8. doi: 10.1111/j.1600-0765.2011.01455.x. Epub 2012 Jan 3. J Periodontal Res. 2012. PMID: 22220967 - Comparative Analyses of Subgingival Microbiome in Chronic Periodontitis Patients with and Without IgA Nephropathy by High Throughput 16S rRNA Sequencing.
Cao Y, Qiao M, Tian Z, Yu Y, Xu B, Lao W, Ma X, Li W. Cao Y, et al. Cell Physiol Biochem. 2018;47(2):774-783. doi: 10.1159/000490029. Epub 2018 May 22. Cell Physiol Biochem. 2018. PMID: 29807361
Cited by
- Subgingival microbiota in health compared to periodontitis and the influence of smoking.
Camelo-Castillo AJ, Mira A, Pico A, Nibali L, Henderson B, Donos N, Tomás I. Camelo-Castillo AJ, et al. Front Microbiol. 2015 Feb 24;6:119. doi: 10.3389/fmicb.2015.00119. eCollection 2015. Front Microbiol. 2015. PMID: 25814980 Free PMC article. - Comparison of three qPCR-based commercial tests for detection of periodontal pathogens.
Van der Weijden F, Rijnen M, Valkenburg C. Van der Weijden F, et al. Sci Rep. 2021 Mar 17;11(1):6141. doi: 10.1038/s41598-021-85305-3. Sci Rep. 2021. PMID: 33731742 Free PMC article. - Metagenomic sequencing reveals altered metabolic pathways in the oral microbiota of sailors during a long sea voyage.
Zheng W, Zhang Z, Liu C, Qiao Y, Zhou D, Qu J, An H, Xiong M, Zhu Z, Zhao X. Zheng W, et al. Sci Rep. 2015 Mar 16;5:9131. doi: 10.1038/srep09131. Sci Rep. 2015. PMID: 26154405 Free PMC article. - Periodontal disease and subgingival microbiota as contributors for rheumatoid arthritis pathogenesis: modifiable risk factors?
Scher JU, Bretz WA, Abramson SB. Scher JU, et al. Curr Opin Rheumatol. 2014 Jul;26(4):424-9. doi: 10.1097/BOR.0000000000000076. Curr Opin Rheumatol. 2014. PMID: 24807405 Free PMC article. Review. - Friends with Benefits: An Inside Look of Periodontal Microbes' Interactions Using Fluorescence In Situ Hybridization-Scoping Review.
Esteves GM, Pereira JA, Azevedo NF, Azevedo AS, Mendes L. Esteves GM, et al. Microorganisms. 2021 Jul 14;9(7):1504. doi: 10.3390/microorganisms9071504. Microorganisms. 2021. PMID: 34361938 Free PMC article.
References
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, et al. (2010) The human oral microbiome. J Bacteriol 192: 5002–5017 JB.00542-10 [pii];10.1128/JB.00542-10 [doi] - DOI - PMC - PubMed
- Duran-Pinedo AE, Paster B, Teles R, Frias-Lopez J (2011) Correlation network analysis applied to complex biofilm communities. PLoS ONE 6: e28438 10.1371/journal.pone.0028438 [doi];PONE-D-11-18472 [pii] - DOI - PMC - PubMed
- Jenkinson HF (2011) Beyond the oral microbiome. Environ Microbiol 13: 3077–3087 10.1111/j.1462-2920.2011.02573.x [doi] - DOI - PubMed
- McLean JS, Fansler SJ, Majors PD, McAteer K, Allen LZ, et al. (2012) Identifying low pH active and lactate-utilizing taxa within oral microbiome communities from healthy children using stable isotope probing techniques. PLoS ONE 7: e32219 10.1371/journal.pone.0032219 [doi];PONE-D-11-21517 [pii] - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources