Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action - PubMed (original) (raw)
Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action
Pengcheng Wang et al. Proc Natl Acad Sci U S A. 2013.
Abstract
Sucrose nonfermenting 1 (SNF1)-related protein kinase 2s (SnRK2s) are central components of abscisic acid (ABA) signaling pathways. The snrk2.2/2.3/2.6 triple-mutant plants are nearly completely insensitive to ABA, suggesting that most of the molecular actions of ABA are triggered by the SnRK2s-mediated phosphorylation of substrate proteins. Only a few substrate proteins of the SnRK2s are known. To identify additional substrate proteins of the SnRK2s and provide insight into the molecular actions of ABA, we used quantitative phosphoproteomics to compare the global changes in phosphopeptides in WT and snrk2.2/2.3/2.6 triple mutant seedlings in response to ABA treatment. Among the 5,386 unique phosphorylated peptides identified in this study, we found that ABA can increase the phosphorylation of 166 peptides and decrease the phosphorylation of 117 peptides in WT seedlings. In the snrk2.2/2.3/2.6 triple mutant, 84 of the 166 peptides, representing 58 proteins, could not be phosphorylated, or phosphorylation was not increased under ABA treatment. In vitro kinase assays suggest that most of the 58 proteins can serve as substrates of the SnRK2s. The SnRK2 substrates include proteins involved in flowering time regulation, RNA and DNA binding, miRNA and epigenetic regulation, signal transduction, chloroplast function, and many other cellular processes. Consistent with the SnRK2 phosphorylation of flowering time regulators, the snrk2.2/2.3/2.6 triple mutant flowered significantly earlier than WT. These results shed new light on the role of the SnRK2 protein kinases and on the downstream effectors of ABA action, and improve our understanding of plant responses to adverse environments.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
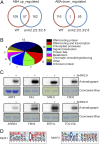
(A) Venn diagram illustrating the overlap of up-regulated (Left) and down-regulated (Right) phosphopeptides identified in WT and snrk2.2/2.3/2.6 triple mutant. (B) GO catalogs of the 58 putative SnRK2 substrates. (C) In vitro phosphorylation of putative substrates by recombinant SnRK2.6. (D) Sequence logos (generated using Motif-X) of the phosphorylation sites seen only in ABA up-regulated phosphopeptides.
Fig. 2.
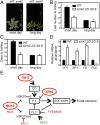
Role of SnRK2.2/2.3/2.6 and ABA up-regulated phosphoproteins in flowering time regulation. (A) Early flowering phenotype of the snrk2.2/2.3/2.6 triple mutant (snrk2) under short-day (Upper; 35-d-old plants) and long-day (Lower; 24-d-old plants) photoperiods. (Scale bars: 10 mm). (B and C) Number of days (B) and vegetative leaf number (C) at bolting of WT and the snrk2.2/2.3/2.6 triple mutant under long-day and short-day photoperiods. Error bars depict SE (n > 25). Asterisks denote significant differences (P < 0.05) between triple-mutant and WT plants. (D) Expression of the flowering time-related genes analyzed by qPCR in leaves of 21-d-old plants in WT and snrk2.2/2.3/2.6 triple mutant under long-day conditions. The experiment was repeated at least twice, with similar results obtained each time. (E) Model showing the function of flowering time-related phosphoproteins identified in this study. The phosphoproteins up-regulated and those down-regulated by ABA treatment are shown in red and green, respectively. Bold letters indicate putative SnRK2 substrates.
Fig. 3.
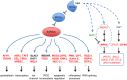
Model showing the regulation and functional grouping of putative SnRK2 substrates in ABA response pathways. The phosphoproteins up-regulated or down-regulated by ABA treatment are shown in red and green, respectively. Bold letters indicate putative SnRK2 substrates.
Similar articles
- Rapid Phosphoproteomic Effects of Abscisic Acid (ABA) on Wild-Type and ABA Receptor-Deficient A. thaliana Mutants.
Minkoff BB, Stecker KE, Sussman MR. Minkoff BB, et al. Mol Cell Proteomics. 2015 May;14(5):1169-82. doi: 10.1074/mcp.M114.043307. Epub 2015 Feb 18. Mol Cell Proteomics. 2015. PMID: 25693798 Free PMC article. - Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana.
Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, Shinozaki K. Umezawa T, et al. Sci Signal. 2013 Apr 9;6(270):rs8. doi: 10.1126/scisignal.2003509. Sci Signal. 2013. PMID: 23572148 - Initiation and amplification of SnRK2 activation in abscisic acid signaling.
Lin Z, Li Y, Wang Y, Liu X, Ma L, Zhang Z, Mu C, Zhang Y, Peng L, Xie S, Song CP, Shi H, Zhu JK, Wang P. Lin Z, et al. Nat Commun. 2021 Apr 28;12(1):2456. doi: 10.1038/s41467-021-22812-x. Nat Commun. 2021. PMID: 33911084 Free PMC article. - The Multifaceted Regulation of SnRK2 Kinases.
Maszkowska J, Szymańska KP, Kasztelan A, Krzywińska E, Sztatelman O, Dobrowolska G. Maszkowska J, et al. Cells. 2021 Aug 24;10(9):2180. doi: 10.3390/cells10092180. Cells. 2021. PMID: 34571829 Free PMC article. Review. - Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants.
Fujita Y, Yoshida T, Yamaguchi-Shinozaki K. Fujita Y, et al. Physiol Plant. 2013 Jan;147(1):15-27. doi: 10.1111/j.1399-3054.2012.01635.x. Epub 2012 May 16. Physiol Plant. 2013. PMID: 22519646 Review.
Cited by
- Abscisic acid regulates stomatal production by imprinting a SnRK2 kinase-mediated phosphocode on the master regulator SPEECHLESS.
Yang X, Gavya S L, Zhou Z, Urano D, Lau OS. Yang X, et al. Sci Adv. 2022 Oct 7;8(40):eadd2063. doi: 10.1126/sciadv.add2063. Epub 2022 Oct 7. Sci Adv. 2022. PMID: 36206348 Free PMC article. - Cellular Phosphorylation Signaling and Gene Expression in Drought Stress Responses: ABA-Dependent and ABA-Independent Regulatory Systems.
Soma F, Takahashi F, Yamaguchi-Shinozaki K, Shinozaki K. Soma F, et al. Plants (Basel). 2021 Apr 13;10(4):756. doi: 10.3390/plants10040756. Plants (Basel). 2021. PMID: 33924307 Free PMC article. Review. - Rapid Phosphoproteomic Effects of Abscisic Acid (ABA) on Wild-Type and ABA Receptor-Deficient A. thaliana Mutants.
Minkoff BB, Stecker KE, Sussman MR. Minkoff BB, et al. Mol Cell Proteomics. 2015 May;14(5):1169-82. doi: 10.1074/mcp.M114.043307. Epub 2015 Feb 18. Mol Cell Proteomics. 2015. PMID: 25693798 Free PMC article. - Distinct guard cell-specific remodeling of chromatin accessibility during abscisic acid- and CO2-dependent stomatal regulation.
Seller CA, Schroeder JI. Seller CA, et al. Proc Natl Acad Sci U S A. 2023 Dec 26;120(52):e2310670120. doi: 10.1073/pnas.2310670120. Epub 2023 Dec 19. Proc Natl Acad Sci U S A. 2023. PMID: 38113262 Free PMC article. - Arabidopsis Raf-Like Kinase Raf10 Is a Regulatory Component of Core ABA Signaling.
Nguyen QTC, Lee SJ, Choi SW, Na YJ, Song MR, Hoang QTN, Sim SY, Kim MS, Kim JI, Soh MS, Kim SY. Nguyen QTC, et al. Mol Cells. 2019 Sep 30;42(9):646-660. doi: 10.14348/molcells.2019.0173. Mol Cells. 2019. PMID: 31480825 Free PMC article.
References
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. - PubMed
- Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324(5930):1064–1068. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases