Mitochondrial fusion proteins and human diseases - PubMed (original) (raw)
Mitochondrial fusion proteins and human diseases
Michela Ranieri et al. Neurol Res Int. 2013.
Abstract
Mitochondria are highly dynamic, complex organelles that continuously alter their shape, ranging between two opposite processes, fission and fusion, in response to several stimuli and the metabolic demands of the cell. Alterations in mitochondrial dynamics due to mutations in proteins involved in the fusion-fission machinery represent an important pathogenic mechanism of human diseases. The most relevant proteins involved in the mitochondrial fusion process are three GTPase dynamin-like proteins: mitofusin 1 (MFN1) and 2 (MFN2), located in the outer mitochondrial membrane, and optic atrophy protein 1 (OPA1), in the inner membrane. An expanding number of degenerative disorders are associated with mutations in the genes encoding MFN2 and OPA1, including Charcot-Marie-Tooth disease type 2A and autosomal dominant optic atrophy. While these disorders can still be considered rare, defective mitochondrial dynamics seem to play a significant role in the molecular and cellular pathogenesis of more common neurodegenerative diseases, for example, Alzheimer's and Parkinson's diseases. This review provides an overview of the basic molecular mechanisms involved in mitochondrial fusion and focuses on the alteration in mitochondrial DNA amount resulting from impairment of mitochondrial dynamics. We also review the literature describing the main disorders associated with the disruption of mitochondrial fusion.
Figures
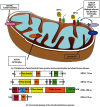
Figure 1
Distribution of mitochondrial fusion proteins inside mitochondria and related human diseases resulting from defects in gene expression or protein function (a). Mitofusin 2 (MFN2) and optic atrophy protein 3 (OPA3) are present in the outer mitochondrial membrane; dynamin-related protein optic atrophy 1 (OPA1) is localized in the intermembrane space and is tethered to the inner mitochondrial membrane. The top right of the figure illustrates the molecular pathway responsible for mitophagy: PTEN-induced putative kinase protein 1 (PINK1) phosphorylates Parkin, an ubiquitin E3 ligase that targets several outer membrane proteins including mitofusin. Ubiquitination of MFN2 in damaged mitochondria starts the mitophagic process. ADOA: autosomal dominant optic atrophy; ADOAC: autosomal dominant optic atrophy and cataract; HR: heptad repeat; PD: Parkinson's disease. Below (b), schematics of functional domains of the mitochondrial fusion proteins (HR: heptad repeat domain; TM: transmembrane domain; PR: proline-rich domain; MIS: mitochondrial import sequence; GED: GTPase effector domain; Mss: mitochondrial signal sequence).
Similar articles
- The role of mitochondrial network dynamics in the pathogenesis of Charcot-Marie-Tooth disease.
Palau F, Estela A, Pla-Martín D, Sánchez-Piris M. Palau F, et al. Adv Exp Med Biol. 2009;652:129-37. doi: 10.1007/978-90-481-2813-6_9. Adv Exp Med Biol. 2009. PMID: 20225023 Review. - Mitochondrial dynamics in cell death and neurodegeneration.
Cho DH, Nakamura T, Lipton SA. Cho DH, et al. Cell Mol Life Sci. 2010 Oct;67(20):3435-47. doi: 10.1007/s00018-010-0435-2. Epub 2010 Jun 25. Cell Mol Life Sci. 2010. PMID: 20577776 Free PMC article. Review. - Mitochondrial dynamics and inherited peripheral nerve diseases.
Pareyson D, Saveri P, Sagnelli A, Piscosquito G. Pareyson D, et al. Neurosci Lett. 2015 Jun 2;596:66-77. doi: 10.1016/j.neulet.2015.04.001. Epub 2015 Apr 3. Neurosci Lett. 2015. PMID: 25847151 Review. - Molecular mechanism of mitochondrial membrane fusion.
Griffin EE, Detmer SA, Chan DC. Griffin EE, et al. Biochim Biophys Acta. 2006 May-Jun;1763(5-6):482-9. doi: 10.1016/j.bbamcr.2006.02.003. Epub 2006 Mar 9. Biochim Biophys Acta. 2006. PMID: 16571363 Review. - Implications of mitochondrial dynamics on neurodegeneration and on hypothalamic dysfunction.
Zorzano A, Claret M. Zorzano A, et al. Front Aging Neurosci. 2015 Jun 10;7:101. doi: 10.3389/fnagi.2015.00101. eCollection 2015. Front Aging Neurosci. 2015. PMID: 26113818 Free PMC article. Review.
Cited by
- Mitochondrial sirtuins and their relationships with metabolic disease and cancer.
Kumar S, Lombard DB. Kumar S, et al. Antioxid Redox Signal. 2015 Apr 20;22(12):1060-77. doi: 10.1089/ars.2014.6213. Epub 2015 Feb 10. Antioxid Redox Signal. 2015. PMID: 25545135 Free PMC article. Review. - Mitochondrial Membranes and Mitochondrial Genome: Interactions and Clinical Syndromes.
Almannai M, Salah A, El-Hattab AW. Almannai M, et al. Membranes (Basel). 2022 Jun 15;12(6):625. doi: 10.3390/membranes12060625. Membranes (Basel). 2022. PMID: 35736332 Free PMC article. Review. - Drp1-mediated mitochondrial fission regulates calcium and F-actin dynamics during wound healing.
Ponte S, Carvalho L, Gagliardi M, Campos I, Oliveira PJ, Jacinto A. Ponte S, et al. Biol Open. 2020 May 3;9(5):bio048629. doi: 10.1242/bio.048629. Biol Open. 2020. PMID: 32184231 Free PMC article. - Mitofusin-2 prevents skeletal muscle wasting in cancer cachexia.
Xi QL, Zhang B, Jiang Y, Zhang HS, Meng QY, Chen Y, Han YS, Zhuang QL, Han J, Wang HY, Fang J, Wu GH. Xi QL, et al. Oncol Lett. 2016 Nov;12(5):4013-4020. doi: 10.3892/ol.2016.5191. Epub 2016 Sep 26. Oncol Lett. 2016. PMID: 27895764 Free PMC article. - Regulation of mitochondrial bioenergetics by the non-canonical roles of mitochondrial dynamics proteins in the heart.
Wang W, Fernandez-Sanz C, Sheu SS. Wang W, et al. Biochim Biophys Acta Mol Basis Dis. 2018 May;1864(5 Pt B):1991-2001. doi: 10.1016/j.bbadis.2017.09.004. Epub 2017 Sep 14. Biochim Biophys Acta Mol Basis Dis. 2018. PMID: 28918113 Free PMC article. Review.
References
- Yoneda M, Miyatake T, Attardi G. Complementation of mutant and wild-type human mitochondrial DNAs coexisting since the mutation event and lack of complementation of DNAs introduced separately into a cell within distinct organelles. Molecular and Cellular Biology. 1994;14(4):2699–2712. - PMC - PubMed
- Nakada K, Inoue K, Ono T, et al. Inter-mitochondrial complementation: mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nature Medicine. 2001;7(8):934–940. - PubMed
- Milone M, Benarroch EE. Mitochondrial dynamics: general concepts and clinical implications. Neurology. 2012;78(20):1612–1619. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials