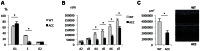BMS1 is mutated in aplasia cutis congenita - PubMed (original) (raw)
BMS1 is mutated in aplasia cutis congenita
Alexander G Marneros. PLoS Genet. 2013 Jun.
Abstract
Aplasia cutis congenita (ACC) manifests with localized skin defects at birth of unknown cause, mostly affecting the scalp vertex. Here, genome-wide linkage analysis and exome sequencing was used to identify the causative mutation in autosomal dominant ACC. A heterozygous Arg-to-His missense mutation (p.R930H) in the ribosomal GTPase BMS1 is identified in ACC that is associated with a delay in 18S rRNA maturation, consistent with a role of BMS1 in processing of pre-rRNAs of the small ribosomal subunit. Mutations that affect ribosomal function can result in a cell cycle defect and ACC skin fibroblasts with the BMS1 p.R930H mutation show a reduced cell proliferation rate due to a p21-mediated G1/S phase transition delay. Unbiased comparative global transcript and proteomic analyses of ACC fibroblasts with this mutation confirm a central role of increased p21 levels for the ACC phenotype, which are associated with downregulation of heterogenous nuclear ribonucleoproteins (hnRNPs) and serine/arginine-rich splicing factors (SRSFs). Functional enrichment analysis of the proteomic data confirmed a defect in RNA post-transcriptional modification as the top-ranked cellular process altered in ACC fibroblasts. The data provide a novel link between BMS1, the cell cycle, and skin morphogenesis.
Conflict of interest statement
The author has declared that no competing interests exist.
Figures
Figure 1. A mutation in the ribosome biogenesis GTPase BMS1 causes autosomal-dominant ACC.
a. Representative images of individuals with ACC in a five-generation pedigree with autosomal dominant inheritance pattern and full penetrance. (Left image: 1 year old with hypertrophic scar at site of congenital ACC of the scalp; Middle image: father of 1 year old of left image with scar at site of congenital ACC; Right image: daughter of this father on day 1 after birth with localized erosion on vertex of scalp (indicated with arrow in pedigree; this member was born after the linkage study was completed and carried the disease allele as well)). Family members' DNA used for genome-wide SNP genotyping indicted with black stars; additional family members' DNA included for confirmation of linkage with microsatellite markers indicated in red stars. b. Histogram of multi-point LOD scores along chromosome 10 shows a single chromosomal region with LOD scores >2 on chromosome 10q11. c. A heterozygous missense mutation resulting in a G>A nucleotide change in the BMS1 gene was identified in all affected members with ACC in this family (c.2789G>A). d. The c.2789G>A mutation results in a Arg-to-His amino acid change (p.R930H) of a conserved Arg residue in BMS1. e. The p.R930H amino acid change affects an Arg residue within the putative C-terminal GAP domain of BMS1. Representative image of the domains of BMS1 with its GTPase domain at the N-terminus (adapted from [9]). f. Murine embryonic sections showing the site of skin formation at the vertex area of the scalp at E13.5 (f. and g.). BMS1 expression is seen in all cells, including the proliferative epidermis (C-terminus of BMS1 in red, pH3b green). C-term indicates labeling with the monoclonal anti-BMS1 antibody that recognizes the C-terminus of BMS1. pH3b indicates staining for the proliferation marker phospho-Histone 3B (Ser10) (pH3b, green). g. Co-localization of BMS1 C-terminal (C-term red) and N-terminal domains (N-term green) at E13.5 of embryonic murine scalp at E13.5. Nuclei are labeled with DAPI. All images are acquired with a 20× objective.
Figure 2. ACC fibroblasts with mutated BMS1 show a delay in 18S pre-rRNA processing.
a. Pre-rRNA processing pathways in human cells (adapted from [33]). Two pathways coexist depending on the order of cleavage in the 5′-ETS (sites A0 and A1) and ITS1 (A2). Nomenclature of the cleavage sites follows Hadjiolova et al. (1993) and Rouquette et al. (2005) , . b. Pulse-chase labeling of pre-rRNAs with L-[_methyl_-3H]methionine shows that ACCBMS1(p.R930H) fibroblasts (ACC) form all pre-rRNA species compared to control fibroblasts (WT), but retain increased 45S and 30S pre-rRNAs (seen at 180 minutes after pulse-labeling). Relative values for bands normalized to 28S rRNA band are shown (fold-change compared to WT). c. Magnification of 45S pre-rRNAs 180 minutes after pulse labeling shows retained 45S pre-rRNA in ACCBMS1(p.R930H) fibroblasts compared to control fibroblasts (WT). d. Northern blot analysis of pre-rRNA processing using a radioactive ITS-1 probe. RNA from ACC fibroblasts was used carrying the BMS1 p.R930H mutation, as well as from control fibroblasts (WT). The Northern blot shown on the left side for the WT and ACC cells was exposed overnight, while the image on the right side shows the same blot after a 4 hour exposure. In addition, RNA from fibroblasts stably transfected with an inducible BMS1 shRNA vector was used, after shRNA-mediated knockdown of BMS1 transcripts was induced by doxycycline (doxy) treatment. Ethidiumbromide stained gels prior to blotting (bottom) confirm equal loading of RNA. Quantitation of band intensity ratios expressed as relative values (fold-change compared to WT or untreated cells). e. Doxycyline-inducible knockdown of BMS1 by shRNA lentivirus (shBMS1). Doxycycline treatment for 48 hours results in ∼60% reduction of BMS1 transcript levels. * P-value<0.05.
Figure 3. ACC fibroblasts with mutated BMS1 show a G1/S phase cell cycle transition delay and reduced cell proliferation rate.
a. Primary ACC fibroblasts show a significantly reduced proportion of cells in S-phase compared to control fibroblasts (WT), consistent with a delay in G1/S phase transition in mutant cells. PI staining and FACS-based analysis. * P-value<0.05 for cells in G1 and S phase. b. Cell proliferation rate is reduced in ACC fibroblasts. * P-value<0.05 for values between d4 to d7. c. In vitro scratch assay of fibroblasts on fibronectin-coated dishes show increased cell migration rate in ACC fibroblasts (assayed at 14 hours after scratch). Images taken with a 2.5× objective. * P-value<0.05. Graph shows area not repopulated with fibroblasts.
Figure 4. An increase in p21 levels in ACC skin fibroblasts.
a. Microarray gene expression profiling experiments (experimental triplicates for ACC vs control fibroblasts) show an upregulation of p21 mRNA levels and a downregulation of SRSF3 mRNA levels in ACC fibroblasts. b. These findings were confirmed by semiquantitative RT-PCR. *P-values<0.05. c. Overexpression of mutant BMS1p.R930H in wild-type fibroblasts phenocopies ACC fibroblasts in showing increased p21 and decreased SRSF3 transcript levels. *P-values<0.05. d. Western blotting experiments show increased p21 protein levels in ACC fibroblasts compared to control fibroblasts, while p53 protein levels are not significantly changed. Relative values for bands normalized to loading control are shown.
Figure 5. Global proteomic analysis in ACC skin fibroblasts.
a. Comparative quantitative proteomic analysis with iTRAQ labeling experiments. Upregulated proteins in ACC fibroblasts are shown in the upper part of the figure and downregulated proteins in the lower part of the figure. The columns show values for the following: Peptides (95%): number of peptides identified with at least 95% confidence; raw p-value; false discovery rate (FDR), fold change of WT vs ACC mean values. b. Network analysis (Ingenuity) identified the most significantly altered networks to include top functions for “RNA-posttranscriptional modification”, “protein synthesis” and “cell cycle” (Fisher exact test –lg p-values indicated). c. Western blotting experiments confirm downregulation of hnRNPA2B1 in ACC fibroblasts compared to control fibroblasts. Relative values for bands normalized to loading control are shown.
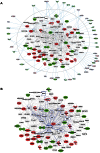
Figure 6. Comparative proteomics and global gene expression profiling reveals a central role of p21 activation in ACC.
a. The two top-ranked networks were merged to generate a combined network of the differentially present proteins in ACC fibroblasts (center). Differentially expressed genes identified in the transcript profiling microarray experiments are indicated in the periphery of the proteomic network, and interactions are illustrated by arrows. This analysis revealed the highest number of interactions to include p21 (CDKN1A) and hnRNPA2B1. b. Interaction analysis among p21 (CDKN1A) and the entire merged proteomic network revealed a central role of p21, showing that p21 activation is a central determinant of the ACC phenotype.
Comment in
- [BMS1 mutations in aplasia cutis congenita].
Dereure O. Dereure O. Ann Dermatol Venereol. 2014 Feb;141(2):161. doi: 10.1016/j.annder.2013.10.034. Epub 2013 Nov 19. Ann Dermatol Venereol. 2014. PMID: 24507215 French. No abstract available.
Similar articles
- [BMS1 mutations in aplasia cutis congenita].
Dereure O. Dereure O. Ann Dermatol Venereol. 2014 Feb;141(2):161. doi: 10.1016/j.annder.2013.10.034. Epub 2013 Nov 19. Ann Dermatol Venereol. 2014. PMID: 24507215 French. No abstract available. - Aplasia Cutis Congenita Pathomechanisms Reveal Key Regulators of Skin and Skin Appendage Morphogenesis.
Marneros AG. Marneros AG. J Invest Dermatol. 2024 Nov;144(11):2399-2405. doi: 10.1016/j.jid.2024.05.014. Epub 2024 Jul 17. J Invest Dermatol. 2024. PMID: 39023472 Free PMC article. Review. - Genetics of aplasia cutis reveal novel regulators of skin morphogenesis.
Marneros AG. Marneros AG. J Invest Dermatol. 2015 Mar;135(3):666-672. doi: 10.1038/jid.2014.413. Epub 2014 Oct 30. J Invest Dermatol. 2015. PMID: 25355129 Review. - Autosomal dominant aplasia cutis in three generations and one case with preaxial polydactyly in the last generation.
Yağci-Küpeli B, Çağlar K, Büyük S, Balci S. Yağci-Küpeli B, et al. Genet Couns. 2011;22(1):55-61. Genet Couns. 2011. PMID: 21614989 - Aplasia cutis congenita in a CDC42-related developmental phenotype.
Schnabel F, Kamphausen SB, Funke R, Kaulfuß S, Wollnik B, Zenker M. Schnabel F, et al. Am J Med Genet A. 2021 Mar;185(3):850-855. doi: 10.1002/ajmg.a.62009. Epub 2020 Dec 7. Am J Med Genet A. 2021. PMID: 33283961
Cited by
- Crucial role of the Rcl1p-Bms1p interaction for yeast pre-ribosomal RNA processing.
Delprato A, Al Kadri Y, Pérébaskine N, Monfoulet C, Henry Y, Henras AK, Fribourg S. Delprato A, et al. Nucleic Acids Res. 2014 Sep;42(15):10161-72. doi: 10.1093/nar/gku682. Epub 2014 Jul 26. Nucleic Acids Res. 2014. PMID: 25064857 Free PMC article. - Membranous aplasia cutis congenita in trisomy 18.
Cammarata-Scalisi F, Diociaiuti A, de Guerrero B, Willoughby CE, Callea M. Cammarata-Scalisi F, et al. Ital J Pediatr. 2020 Aug 27;46(1):120. doi: 10.1186/s13052-020-00885-6. Ital J Pediatr. 2020. PMID: 32854736 Free PMC article. - Aplasia cutis congenita: a report of two cases from National Hospital Abuja, Nigeria and review of the literature.
Mukhtar-Yola M, Mshelia L, Mairami AB, Otuneye AT, Yawe ET, Igoche P, Audu LI. Mukhtar-Yola M, et al. Pan Afr Med J. 2020 Aug 17;36:291. doi: 10.11604/pamj.2020.36.291.24523. eCollection 2020. Pan Afr Med J. 2020. PMID: 33117485 Free PMC article. Review. - How Altered Ribosome Production Can Cause or Contribute to Human Disease: The Spectrum of Ribosomopathies.
Venturi G, Montanaro L. Venturi G, et al. Cells. 2020 Oct 15;9(10):2300. doi: 10.3390/cells9102300. Cells. 2020. PMID: 33076379 Free PMC article. Review. - Aplasia cutis congenita type VII of the lower extremity: a favourable disease course with minimal conservative treatment.
Quach KT, Wind C, van Mierlo K, Vos LE. Quach KT, et al. BMJ Case Rep. 2024 Apr 17;17(4):e257572. doi: 10.1136/bcr-2023-257572. BMJ Case Rep. 2024. PMID: 38631814 Free PMC article.
References
- Evers ME, Steijlen PM, Hamel BC (1995) Aplasia cutis congenita and associated disorders: an update. Clin Genet 47: 295–301. - PubMed
- Zenker M, Mayerle J, Lerch MM, Tagariello A, Zerres K, et al. (2005) Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome). Nat Genet 37: 1345–1350. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases