Uniparental genetic heritage of belarusians: encounter of rare middle eastern matrilineages with a central European mitochondrial DNA pool - PubMed (original) (raw)
. 2013 Jun 13;8(6):e66499.
doi: 10.1371/journal.pone.0066499. Print 2013.
Larysa Sivitskaya, Nina Danilenko, Tadeush Novogrodskii, Iosif Tsybovsky, Anna Kiseleva, Svetlana Kotova, Gyaneshwer Chaubey, Ene Metspalu, Hovhannes Sahakyan, Ardeshir Bahmanimehr, Maere Reidla, Siiri Rootsi, Jüri Parik, Tuuli Reisberg, Alessandro Achilli, Baharak Hooshiar Kashani, Francesca Gandini, Anna Olivieri, Doron M Behar, Antonio Torroni, Oleg Davydenko, Richard Villems
Affiliations
- PMID: 23785503
- PMCID: PMC3681942
- DOI: 10.1371/journal.pone.0066499
Uniparental genetic heritage of belarusians: encounter of rare middle eastern matrilineages with a central European mitochondrial DNA pool
Alena Kushniarevich et al. PLoS One. 2013.
Abstract
Ethnic Belarusians make up more than 80% of the nine and half million people inhabiting the Republic of Belarus. Belarusians together with Ukrainians and Russians represent the East Slavic linguistic group, largest both in numbers and territory, inhabiting East Europe alongside Baltic-, Finno-Permic- and Turkic-speaking people. Till date, only a limited number of low resolution genetic studies have been performed on this population. Therefore, with the phylogeographic analysis of 565 Y-chromosomes and 267 mitochondrial DNAs from six well covered geographic sub-regions of Belarus we strove to complement the existing genetic profile of eastern Europeans. Our results reveal that around 80% of the paternal Belarusian gene pool is composed of R1a, I2a and N1c Y-chromosome haplogroups - a profile which is very similar to the two other eastern European populations - Ukrainians and Russians. The maternal Belarusian gene pool encompasses a full range of West Eurasian haplogroups and agrees well with the genetic structure of central-east European populations. Our data attest that latitudinal gradients characterize the variation of the uniparentally transmitted gene pools of modern Belarusians. In particular, the Y-chromosome reflects movements of people in central-east Europe, starting probably as early as the beginning of the Holocene. Furthermore, the matrilineal legacy of Belarusians retains two rare mitochondrial DNA haplogroups, N1a3 and N3, whose phylogeographies were explored in detail after de novo sequencing of 20 and 13 complete mitogenomes, respectively, from all over Eurasia. Our phylogeographic analyses reveal that two mitochondrial DNA lineages, N3 and N1a3, both of Middle Eastern origin, might mark distinct events of matrilineal gene flow to Europe: during the mid-Holocene period and around the Pleistocene-Holocene transition, respectively.
Conflict of interest statement
Competing Interests: Co-authors Alessandro Achilli and Doron M Behar are Academic Editors for PLOS ONE. This does not alter the authors' adherence to all the PLOS ONE policies on sharing data and materials.
Figures
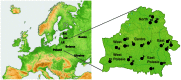
Figure 1. Geographic position of Belarus within Europe.
Map of Belarus demonstrating the six geographic sub-regions studied is shown on the right. Numbers 1–19 correspond to the location of sampling points (Table S1). Lit – Lithuania, Lat – Latvia, Est – Estonia.
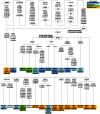
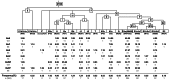
Figure 2. Phylogeny of mtDNA haplogroups and their relative frequencies in Belarusians.
The tree is rooted relative to the RSRS according to . Belarusian sub-populations are designated as BeE – East, BeWP – West Polesie, BeEP – East Polesie, BeN – North, BeC – Centre, BeW – West. Sample sizes and absolute frequencies are also given.
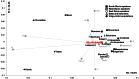
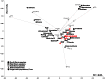
Figure 3. PC analysis based on mtDNA haplogroup frequencies among eastern Europeans and Balkan populations.
The contribution of each haplogroup to the first and the second PCs is shown in gray. The group “Other” includes “Other” from published data merged with uncommon haplogroups L1b, L2a and L3f. Frequencies of mtDNA haplogroups and references are listed in Table S3.
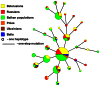
Figure 4. Maximum parsimony tree of mtDNA haplogroup N1a3.
The tree includes 20 novel complete sequences (marked with an asterisk and underlined accession numbers) and eight previously published , [50(and references therein)], . Mutations relative to the RSRS are indicated on the branches; transversions are specified with a lower case letter; Y and R stand for heteroplasmy; underlining indicates positions experiencing recurrent mutations within the tree while exclamation marks refer to one (!) or two (!!) back mutations relative to the RSRS. Coalescence age estimates for N1a3 and N1a3a obtained by employing the complete genome and synonymous (ρ) clocks, indicated by # and @, respectively, are also shown.
Figure 5. Maximum parsimony tree of mtDNA haplogroup N3.
The tree includes 13 novel (marked with an asterisk and underlined accession numbers) and three previously published , complete sequences. Mutations relative to the RSRS are shown on the branches; transversions are specified with a lower case letter; underlining indicates positions which experienced recurrent mutations within the tree, while the exclamation mark (!) refers to one back mutation relative to the RSRS. Rho coalescence time estimates and their confidence intervals for haplogroup N3 and its major sub-branch N3a obtained from the complete genome clock are also shown.
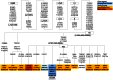
Figure 6. Phylogeny of NRY haplogroups and their relative frequencies in Belarusians.
Haplogroup-defining biallelic markers are in parentheses. Belarusian sub-populations are designated as BeN – North, BeC – Centre, BeE – East, BeW – West, BeWP – West Polesie, BeEP – East Polesie. Sample sizes and absolute frequencies are also given.
Figure 7. Maximum Parsimony tree (based on MJ network) of NRY haplogroup N1c(Tat) calculated from seven Y-STRs.
Volga-Uralic populations include Komis (Priluzhski, Izhevski), Udmurts, Maris, Bashkirs, Chuvashes. Altogether 402 individuals are analyzed, the sample size of each population and the set of Y-STRs used for calculations are given in Table S9.
Figure 8. Maximum Parsimony tree (based on MJ network) of NRY haplogroup I2a(P37) calculated from seven Y-STRs.
Balkan populations include Bosnians, Croatians and Slovenians. Altogether 347 individuals are analyzed, the sample size of each population and the set of Y-STRs used for calculations are given in Table S9.
Figure 9. PC plot based on NRY haplogroup frequencies among eastern Europeans and Balkan populations.
The contribution of each haplogroup to the first and the second PCs are shown in gray. Population abbreviations are as follows: BeN, BeW, BeC, BeWP, BeEP, BeE – Belarusians from North, West, Central, West Polesie, East Polesie and East sub-regions, respectively, filled red circle denotes the total Belarusian population; RuS, RuC, RuN – Russians from southern, central and northern regions, respectively; Finns K – Finns from Karelia. K*(x N,P) refers to samples with M9, M20, M70 derived alleles and 92R7, M214 ancestral alleles; P*(xR) refers to samples with 92R7, M242 derived alleles and M207 ancestral allele; F*(xI,J,K) refers to samples with M89 derived allele and M9, M201, M170, 12f2 ancestral alleles; C(xF)DE refers to samples with Yap and M130 derived and M89 ancestral alleles. Frequencies of NRY haplogroups and references are listed in Table S3.
Similar articles
- Autosomal and uniparental portraits of the native populations of Sakha (Yakutia): implications for the peopling of Northeast Eurasia.
Fedorova SA, Reidla M, Metspalu E, Metspalu M, Rootsi S, Tambets K, Trofimova N, Zhadanov SI, Hooshiar Kashani B, Olivieri A, Voevoda MI, Osipova LP, Platonov FA, Tomsky MI, Khusnutdinova EK, Torroni A, Villems R. Fedorova SA, et al. BMC Evol Biol. 2013 Jun 19;13:127. doi: 10.1186/1471-2148-13-127. BMC Evol Biol. 2013. PMID: 23782551 Free PMC article. - Complete mitochondrial DNA analysis of eastern Eurasian haplogroups rarely found in populations of northern Asia and eastern Europe.
Derenko M, Malyarchuk B, Denisova G, Perkova M, Rogalla U, Grzybowski T, Khusnutdinova E, Dambueva I, Zakharov I. Derenko M, et al. PLoS One. 2012;7(2):e32179. doi: 10.1371/journal.pone.0032179. Epub 2012 Feb 21. PLoS One. 2012. PMID: 22363811 Free PMC article. - High-resolution phylogenetic analysis of southeastern Europe traces major episodes of paternal gene flow among Slavic populations.
Pericić M, Lauc LB, Klarić IM, Rootsi S, Janićijevic B, Rudan I, Terzić R, Colak I, Kvesić A, Popović D, Sijacki A, Behluli I, Dordevic D, Efremovska L, Bajec DD, Stefanović BD, Villems R, Rudan P. Pericić M, et al. Mol Biol Evol. 2005 Oct;22(10):1964-75. doi: 10.1093/molbev/msi185. Epub 2005 Jun 8. Mol Biol Evol. 2005. PMID: 15944443 - Phylogeographic review of Y chromosome haplogroups in Europe.
Navarro-López B, Granizo-Rodríguez E, Palencia-Madrid L, Raffone C, Baeta M, de Pancorbo MM. Navarro-López B, et al. Int J Legal Med. 2021 Sep;135(5):1675-1684. doi: 10.1007/s00414-021-02644-6. Epub 2021 Jul 3. Int J Legal Med. 2021. PMID: 34216266 Review. - Review of Croatian genetic heritage as revealed by mitochondrial DNA and Y chromosomal lineages.
Pericić M, Barać Lauc L, Martinović Klarić I, Janićijević B, Rudan P. Pericić M, et al. Croat Med J. 2005 Aug;46(4):502-13. Croat Med J. 2005. PMID: 16100752 Review.
Cited by
- East Eurasian ancestry in the middle of Europe: genetic footprints of Steppe nomads in the genomes of Belarusian Lipka Tatars.
Pankratov V, Litvinov S, Kassian A, Shulhin D, Tchebotarev L, Yunusbayev B, Möls M, Sahakyan H, Yepiskoposyan L, Rootsi S, Metspalu E, Golubenko M, Ekomasova N, Akhatova F, Khusnutdinova E, Heyer E, Endicott P, Derenko M, Malyarchuk B, Metspalu M, Davydenko O, Villems R, Kushniarevich A. Pankratov V, et al. Sci Rep. 2016 Jul 25;6:30197. doi: 10.1038/srep30197. Sci Rep. 2016. PMID: 27453128 Free PMC article. - Transferability of the PRS estimates for height and BMI obtained from the European ethnic groups to the Western Russian populations.
Albert EA, Kondratieva OA, Baranova EE, Sagaydak OV, Belenikin MS, Zobkova GY, Kuznetsova ES, Deviatkin AA, Zhurov AA, Karpulevich EA, Volchkov PY, Vorontsova MV. Albert EA, et al. Front Genet. 2023 Jan 16;14:1086709. doi: 10.3389/fgene.2023.1086709. eCollection 2023. Front Genet. 2023. PMID: 36726807 Free PMC article. - Mitochondrial ancestry of medieval individuals carelessly interred in a multiple burial from southeastern Romania.
Rusu I, Modi A, Radu C, Mircea C, Vulpoi A, Dobrinescu C, Bodolică V, Potârniche T, Popescu O, Caramelli D, Kelemen B. Rusu I, et al. Sci Rep. 2019 Jan 30;9(1):961. doi: 10.1038/s41598-018-37760-8. Sci Rep. 2019. PMID: 30700787 Free PMC article. - Genetic Heritage of the Balto-Slavic Speaking Populations: A Synthesis of Autosomal, Mitochondrial and Y-Chromosomal Data.
Kushniarevich A, Utevska O, Chuhryaeva M, Agdzhoyan A, Dibirova K, Uktveryte I, Möls M, Mulahasanovic L, Pshenichnov A, Frolova S, Shanko A, Metspalu E, Reidla M, Tambets K, Tamm E, Koshel S, Zaporozhchenko V, Atramentova L, Kučinskas V, Davydenko O, Goncharova O, Evseeva I, Churnosov M, Pocheshchova E, Yunusbayev B, Khusnutdinova E, Marjanović D, Rudan P, Rootsi S, Yankovsky N, Endicott P, Kassian A, Dybo A; Genographic Consortium; Tyler-Smith C, Balanovska E, Metspalu M, Kivisild T, Villems R, Balanovsky O. Kushniarevich A, et al. PLoS One. 2015 Sep 2;10(9):e0135820. doi: 10.1371/journal.pone.0135820. eCollection 2015. PLoS One. 2015. PMID: 26332464 Free PMC article. - Analysis of Y chromosome haplogroups in Parkinson's disease.
Grenn FP, Makarious MB, Bandres-Ciga S, Iwaki H, Singleton AB, Nalls MA, Blauwendraat C; International Parkinson Disease Genomics Consortium (IPDGC). Grenn FP, et al. Brain Commun. 2022 Oct 28;4(6):fcac277. doi: 10.1093/braincomms/fcac277. eCollection 2022. Brain Commun. 2022. PMID: 36387750 Free PMC article.
References
- Mahnach AS, Garetski RG, Matveev AV (2001) Geologiya Belarusi. Minsk: IGN NAN Belarusi. 815 p. (in Russian)
- Svezhentsev YS, Popov SG (1993) Late Paleolithic chronology of the East European Plain. Radiocarbon 35: 495–501.
- Zaikouski EM, Isaenka UF, Kalechyts AG, Kapycin VF, Kryvaltsevich MM (1997) Arhealogiya Belarusi. Minsk: Belaruskaya Navuka. Vol. I. pp. 21–88. (in Belarusian)
- Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, et al. (2000) The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 290: 1155–1159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This study was supported by the State Committee on Science and Technology of the Republic of Belarus (SCST), Progetti Ricerca Interesse Nazionale 2009 (Italian Ministry of the University) (to AA and AT), FIRB-Futuro in Ricerca 2008 (Italian Ministry of the University) (to AA and AO), Fondazione Alma Mater Ticinensis (to AT), Estonian Basic Research grant SF0182474 (to RV and EM), Estonian Science Foundation grants (7858) (to EM), European Commission grant (ECOGENE205419) (to RV), the European Union Regional Development Fund (through the Centre of Excellence in Genomics (to RV). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources