Recombinant probes for visualizing endogenous synaptic proteins in living neurons - PubMed (original) (raw)
. 2013 Jun 19;78(6):971-85.
doi: 10.1016/j.neuron.2013.04.017.
Jason A Junge, Rudy J Mora, Hyung-Bae Kwon, C Anders Olson, Terry T Takahashi, Emily R Liman, Graham C R Ellis-Davies, Aaron W McGee, Bernardo L Sabatini, Richard W Roberts, Don B Arnold
Affiliations
- PMID: 23791193
- PMCID: PMC3779638
- DOI: 10.1016/j.neuron.2013.04.017
Recombinant probes for visualizing endogenous synaptic proteins in living neurons
Garrett G Gross et al. Neuron. 2013.
Abstract
The ability to visualize endogenous proteins in living neurons provides a powerful means to interrogate neuronal structure and function. Here we generate recombinant antibody-like proteins, termed Fibronectin intrabodies generated with mRNA display (FingRs), that bind endogenous neuronal proteins PSD-95 and Gephyrin with high affinity and that, when fused to GFP, allow excitatory and inhibitory synapses to be visualized in living neurons. Design of the FingR incorporates a transcriptional regulation system that ties FingR expression to the level of the target and reduces background fluorescence. In dissociated neurons and brain slices, FingRs generated against PSD-95 and Gephyrin did not affect the expression patterns of their endogenous target proteins or the number or strength of synapses. Together, our data indicate that PSD-95 and Gephyrin FingRs can report the localization and amount of endogenous synaptic proteins in living neurons and thus may be used to study changes in synaptic strength in vivo.
Copyright © 2013 Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest statement.
One of us (GCRE-D) has filed a preliminary patent declaration on the synthesis of dinitroindolinyl-caged neurotransmitters.
Figures
Figure 1
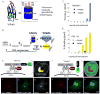
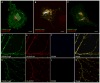
Selection of Fibronectin binders of PSD-95 and Gephyrin by mRNA display and by a cellular localization assay. (A) A library consisting of 10FnIII domains with 17 random residues in the BC and FG loops was used to select binders to PSD-95 and Gephyrin. (B) The selection protocol is as follows: 1. DNA encoding the randomized Fibronectins was transcribed and a puromycin molecule attached to linker DNA was fused to the 3′ end of the transcript. 2. The mRNA-puromycin fusion was translated to give an mRNA-puromycin-peptide molecule. An anti-sense cDNA strand hybridized to the mRNA was synthesized that allows individual library members to be amplified by PCR. 3. The library was exposed to target molecules consisting of either the G domain of Gephyrin or the SH3-GK domains of PSD-95. Binders were purified by precipitation. 4. Library members that bound were amplified by PCR to reconstitute a library that is enriched for binders to target. (C, D) Percentage of the library that bound to the beads after each round of selection. (E) Schematic of a COS cell expressing (1) a FingR that binds to its target with high affinity (winner) and (2) the target domain from the selection (GPHN:G) fused to a Golgi targeting sequence (GTS) and Streptavidin (SA), and tagged with biotin-Rhodamine (Rhod). If the FingR-GFP binds to the target domain with high affinity and specificity, it becomes localized to the Golgi Apparatus and colocalized with GTS-GPHN:G. (F) Expression pattern of GTS-GPHN:G tagged with Rhodamine (red) colocalizes with that of GPHN.FingR.W-GFP (green, G). (H) Yellow indicates colocalization of GTS-GPHN:G and GPHN.FingR.W-GFP. (I) Schematic of COS cell expressing GTS-GPHN:G and a FingR that does not bind to target with high affinitiy and specificity, GPHN.FingR.L-GFP. (J) Expression pattern of GTS-GPHN:G tagged with Rhodamine (red) does not colocalize with that of GPHN.FingR.L-GFP, but instead localizes diffusely (green, K). (L) A relative lack of yellow staining in merge of GTS-GPHN:G and GPHN.FingR.L-GFP indicates a lack of colocalization. Scale bar 5 μm.
Figure 2
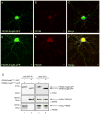
FingRs recognizing Gephyrin or PSD-95 bind to endogenous targets following expression in neurons. (A) A FingR against Gephyrin (GPHN.FingR-GFP, green) localizes in a punctate fashion following expression in a dissociated cortical neuron. (B) Endogenous Gephyrin (red). (C) Merge of GPHN.FingR-GFP and endogenous Gephyrin shows colocalization (yellow). (D) A FingR against PSD-95 (PSD95.FingR-GFP, green) localizes in a punctate fashion following expression in a cortical neuron. (E) Endogenous PSD-95 (red). (F) Merge of PSD95.FingR-GFP and endogenous PSD-95 shows colocalization (yellow). Scale bar is 5 μm. (G) Co-immunoprecipitation of endogenous target proteins with FingRs following expression in cortical neurons in culture. Neurons were infected with lentivirus expressing either PSD95.FingR(FLAG)-GFP (lanes 1, 3) or GPHN.FingR(FLAG)-GFP (lanes 2, 4), both of which contained a FLAG-tag that enabled immunolabeling. Following 96 hours of expression cells were lysed and a portion of the lysates were exposed to a bead-linked anti-GFP antibody. The resulting precipitates were run on an SDS PAGE gel (lanes 1, 2) along with the lysates (lanes 3, 4), blotted and probed with anti-FLAG antibodies (top), anti-Gephyrin antibodies (middle) and anti-PSD-95 antibodies (bottom). PSD95.FingR(FLAG)-GFP co-immunoprecipitated with endogenous PSD95, but not with endogenous Gephyrin, whereas GPHN.FingR(FLAG)-GFP co-immunoprecipitated with Gephyrin, but not with PSD95. See also Figure S1.
Figure 3
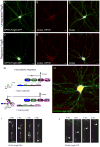
A transcriptional control system causes FingRs to be expressed at the same level as the endogenous target protein. (A) GPHN.FingR-GFP (green) expressed for 7 days localizes in a nonspecific pattern, likely due to high background from unbound FingR. (B, C) Endogenous Gephyrin (red) does not colocalize with GPHN.FingR-GFP. (D) GPHN.FingR-GFP (green) with transcriptional regulation expressed for 7 days appears in a punctate pattern that precisely colocalizes with that of endogenous Gephyrin (red, E, F). Note that red staining that does not colocalize in (F) is from untransfected cells. Scale bar is 5 μm. (G) To control its transcription the FingR (blue) is fused to a transcription factor consisting of a Zinc-Finger DNA binding domain (ZF, purple) fused with a KRAB(A) transcriptional repressor domain (pink). In addition, a ZF DNA binding site (light blue) was inserted upstream of the CMV promoter. When less than 100% of the target is bound by FingR, then 100% of the FingR binds to endogenous target and is prevented from moving to the nucleus. When 100% of the target is bound, then the FingR can no longer bind to the target, and, instead, moves to the nucleus as a result of the nuclear localization signal within the ZF. Once in the nucleus the transcription factor binds to the ZF binding site and represses transcription. Thus, the level of FingR is matched to the level of the endogenous target protein. (H) Live cortical neuron co-expressing regulated PSD95.FingR-GFP (green) and GPHN.FingR-mKate2 (red). Note that the regulation systems for PSD95.FingR-GFP and GPHN.FingR-mKate2 are based on DNA binding domains from CCR5 and IL2RG, respectively. Each FingR is expressed in a punctate, nonoverlapping fashion consistent with labeling of PSD-95 and Gephyrin. (I) Neuron in cortical culture expressing transcriptionally controlled GPHN.FingR-GFP following lentiviral infection. Images taken at 1 second intervals show a vesicle moving with a velocity of ~7 μm.s−1 in the axon. (J) Similarly labeled vesicle moving at ~4 μm.s−1 in the axon. Scale bar is 5 μm. See also Figure S2, Movie S1.
Figure 4
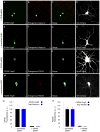
Knock down of Gephyrin or PSD-95 in dissociated cortical neurons leads to elimination of GPHN.FingR and PSD95.FingR staining. (A) Cortical neuron in dissociated culture co-expressing siRNA against Gephyrin and the transcriptionally controlled GPHN.FingR. Note that virtually no staining against the GPHN.FingR is visible in the dendrites or axon except for a single punctum (arrowhead), which colocalizes with a punctum of endogenous Gephyrin (B, C). (D) HA-mCherry staining of the transfected cell. In contrast, when the transcriptionally regulated GPHN.FingR was coexpressed with scrambled siRNA it expressed abundantly in a punctate pattern (E) that colocalized with endogenous Gephyrin (F, G). (H) HA-mCherry staining of the transfected cell. (I) Neuron expressing transcriptionally controlled PSD95.FingR and siRNA shows a very low level of FingR staining that is comparable to the level of endogenous PSD-95 staining (J, K). (L) HA-mCherry staining of transfected cell. In contrast, staining of the transcriptionally regulated PSD95.FingR is robust in a cell coexpressing scrambled siRNA (M) and colocalizes with staining of endogenous PSD-95 (N, O). (P) HA-mCherry staining of transfected cell. Scale bar is 5 μm. (Q) Quantitative comparison of the total amount of staining by anti-Gephyrin antibody (total Gephyrin) and by GPHN.FingR-GFP in the processes of neurons transfected with siRNA against Gephyrin vs. with scrambled siRNA. Note that staining by anti-Gephyrin antibody and by GPHN.FingR-GFP are reduced by comparable amounts in cells expressing Gephyrin siRNA vs. scrambled siRNA. (R) Quantitative comparison of the total amount of staining by anti-PSD-95 antibody (total PSD95) and by PSD95.FingR-GFP in the processes of neurons transfected with siRNA against PSD-95 vs. with scrambled siRNA. Note that staining by anti-PSD-95 antibody and by PSD95.FingR-GFP are reduced by comparable amounts in cells expressing PSD-95 siRNA vs. scrambled siRNA. Merge. See also Figure S3.
Figure 5
Interaction of PSD95.FingR with MAGUK proteins. (A) In a COS cell coexpressing Golgi-targeted PSD-93 (GTS-PSD-93) and PSD95.FingR, the two proteins do not colocalize, indicating the PSD95.FingR does not interact with PSD-93. In contrast, PSD95.FingR colocalizes with GTS-SAP-102 (B) or GTS-SAP-97 (C) when it is co-expressed either MAGUK protein. (D) PSD95.FingR colocalizes with exogenous HA-SAP-102 (E, G) following expression in dissociated cortical neurons where PSD-95 (F) has been knocked down with siRNA. PSD95.FingR (H) colocalizes with exogenous HA-SAP-97 (I, K) following expression in dissociated cortical neurons where PSD-95 (J) has been knocked down with siRNA. Scale bar is 5 μm.
Figure 6
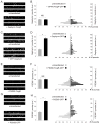
Expression of FingRs does not change the size of Gephyrin or PSD-95 puncta. (A) Immunostained Gephyrin puncta from untransfected cells and cells transfected with GPHN.FingR-GFP are of similar size and brightness. (B) The average intensity associated with Gephyrin puncta is not significantly different in neurons expressing GPHN.FingR-GFP vs. untransfected neurons (p > 0.5). The amounts of Gephyrin / puncta are distributed similarly in neurons expressing GPHN.FingR-GFP vs. untransfected neurons. (C) Puncta stained with an anti-Gephyrin antibody from cells transfected with Gephyrin-GFP tend to be brighter and larger than those from untransfected cells. (D) The average intensity associated with Gephyrin puncta is significantly larger in neurons expressing Gephyrin-GFP vs. untransfected neurons (p < 0.0001). In cells expressing Gephyrin-GFP the amount of Gephyrin / puncta is distributed over a larger range encompassing higher values as compared with similar measurements in untransfected cells. Note that vertical axes of the histograms in (**B**) and (**D**) have the same scale. (**E**) Puncta stained with an anti-PSD-95 antibody from untransfected cells and cells transfected with PSD95.FingR-GFP are of similar size and brightness. (**F**) The average intensity associated with PSD-95 puncta is not significantly different in neurons expressing PSD95.FingR-GFP vs. untransfected neurons (p > 0.5). The amounts of PSD-95 / puncta are distributed similarly in neurons expressing PSD95.FingR-GFP vs. untransfected neurons. (G) Puncta stained with an anti-PSD-95 antibody from cells transfected with PSD95-GFP tend to be brighter and larger than those from untransfected cells. (H) The average intensity associated with PSD-95 puncta is significantly larger in neurons expressing PSD95-GFP vs. untransfected neurons (p < 0.0001). In cells expressing PSD95-GFP the amount of PSD-95 / puncta is distributed over a larger range encompassing higher values as compared with similar measurements in untransfected cells. Note that vertical axes of the histograms in (F) and (H) have the same scale. See also Figure S4.
Figure 7
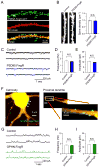
The presence of FingRs does not change the morphology or electrophysiological properties of neurons in hippocampal slices. (A) PSD95.FingR-GFP (green) expressed in CA1 neurons of the hippocampus shows punctate staining of spine heads, consistent with staining at postsynaptic sites, which is distinct from the pattern of coexpressed TdTomato (red). (B) The morphology of dendrites in cells expressing PSD95.FingR-GFP was not qualitatively different from control cells. Similarly, spine density was not significantly different between cells expressing PSD95.FingR-GFP and control cells. (C) mEPSCs from cells expressing PSD95.FingR-GFP were not qualitatively different from those recorded from control cells. In addition the frequencies (D) and amplitudes (E) of mEPSCs did not differ between cells expressing PSD95.FingR-GFP and control cells. (F) In a cell expressing GPHN.FingR-GFP (green) and TdTomato (red) the FingR expresses in puncta on the dendritic shaft in a manner similar to inhibitory postsynaptic sites. (G) mIPSCs recorded from cells expressing GPHN.FingR-GFP do not differ qualitatively from mIPSCs from untransfected control neurons. (H, I) Similarly, neither the frequency nor the amplitude of mIPSCs recorded from cells expressing GPHN.FingR-GFP differed significantly from those recorded from control cells.
Figure 8
Puncta of GPHN.FingR-GFP intrabody recognized by GFP fluorescence represent clusters of ionotropic GABA receptors. (A) Apical dendrite of CA1 pyramidal neurons in an oganotypic slice culture showing the locations of serial GABA uncaging by 2-photon mediated photorelease from CDNI-GABA. (B, C) Resulting inward currents reveal a rapid fall off in the amplitude of IPSCs with distance from the GFP punctum. (D) Lower magnification image of additional dendrite with several labeled GFP puncta and dendritic spines. (E) Representative traces during GABA photorelease at two locations (2, 3) that were not near GPHN.FingR-GFP puncta (one on a dendritic spine and a second on the shaft) and at two locations (1, 4) next to GPHN.FingR-GFP puncta located on the dendritic shaft. The currents are blocked by picrotoxin, as expected for ionotropic GABA receptors.
Figure 9
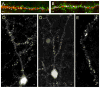
Expression of PSD95.FingR-GFP in vivo. (A, B) Apical dendrites from a layer 5 cortical pyramidal neuron expressing PSD95.FingR-GFP (green) and HAmCherry (red). Cells were present in fixed sections cut from 7 week old mice that were electroporated in utero. (C) Layer 5 cortical pyramidal neuron expressing PSD95.FingR-GFP from section prepared in the same manner as in (A, B). (D) Layer 2/3 cortical neuron expressing PSD95.FingR-GFP from section prepared in the same manner as in (A, B). (E) Live two photon image of a tuft of an apical dendrite from a cortical pyramidal neuron. Image was taken through a cranial window in a 7 week old mouse expressing PSD95.FingR-GFP. Scale bar 5 μm.
Comment in
- Protein GPS.
Pastrana E. Pastrana E. Nat Methods. 2013 Aug;10(8):696-7. doi: 10.1038/nmeth.2588. Nat Methods. 2013. PMID: 24058978 No abstract available.
Similar articles
- Protein GPS.
Pastrana E. Pastrana E. Nat Methods. 2013 Aug;10(8):696-7. doi: 10.1038/nmeth.2588. Nat Methods. 2013. PMID: 24058978 No abstract available. - Gephyrin-Lacking PV Synapses on Neocortical Pyramidal Neurons.
Kuljis DA, Micheva KD, Ray A, Wegner W, Bowman R, Madison DV, Willig KI, Barth AL. Kuljis DA, et al. Int J Mol Sci. 2021 Sep 17;22(18):10032. doi: 10.3390/ijms221810032. Int J Mol Sci. 2021. PMID: 34576197 Free PMC article. - Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin.
Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Naisbitt S, et al. Neuron. 1999 Jul;23(3):569-82. doi: 10.1016/s0896-6273(00)80809-0. Neuron. 1999. PMID: 10433268 - Structural plasticity with preserved topology in the postsynaptic protein network.
Blanpied TA, Kerr JM, Ehlers MD. Blanpied TA, et al. Proc Natl Acad Sci U S A. 2008 Aug 26;105(34):12587-92. doi: 10.1073/pnas.0711669105. Epub 2008 Aug 22. Proc Natl Acad Sci U S A. 2008. PMID: 18723686 Free PMC article. - Postsynaptic scaffolding molecules modulate the localization of neuroligins.
Levinson JN, Li R, Kang R, Moukhles H, El-Husseini A, Bamji SX. Levinson JN, et al. Neuroscience. 2010 Feb 3;165(3):782-93. doi: 10.1016/j.neuroscience.2009.11.016. Epub 2009 Nov 13. Neuroscience. 2010. PMID: 19914352
Cited by
- In vivo super-resolution of the brain - How to visualize the hidden nanoplasticity?
Willig KI. Willig KI. iScience. 2022 Aug 17;25(9):104961. doi: 10.1016/j.isci.2022.104961. eCollection 2022 Sep 16. iScience. 2022. PMID: 36093060 Free PMC article. Review. - Propofol attenuates kinesin-mediated axonal vesicle transport and fusion.
Frank M, Nabb AT, Gilbert SP, Bentley M. Frank M, et al. Mol Biol Cell. 2022 Nov 1;33(13):ar119. doi: 10.1091/mbc.E22-07-0276. Epub 2022 Sep 14. Mol Biol Cell. 2022. PMID: 36103253 Free PMC article. - Relocation of an Extrasynaptic GABAA Receptor to Inhibitory Synapses Freezes Excitatory Synaptic Strength and Preserves Memory.
Davenport CM, Rajappa R, Katchan L, Taylor CR, Tsai MC, Smith CM, de Jong JW, Arnold DB, Lammel S, Kramer RH. Davenport CM, et al. Neuron. 2021 Jan 6;109(1):123-134.e4. doi: 10.1016/j.neuron.2020.09.037. Epub 2020 Oct 22. Neuron. 2021. PMID: 33096025 Free PMC article. - Distinct in vivo dynamics of excitatory synapses onto cortical pyramidal neurons and parvalbumin-positive interneurons.
Melander JB, Nayebi A, Jongbloets BC, Fortin DA, Qin M, Ganguli S, Mao T, Zhong H. Melander JB, et al. Cell Rep. 2021 Nov 9;37(6):109972. doi: 10.1016/j.celrep.2021.109972. Cell Rep. 2021. PMID: 34758304 Free PMC article. - Gephyrin: a master regulator of neuronal function?
Tyagarajan SK, Fritschy JM. Tyagarajan SK, et al. Nat Rev Neurosci. 2014 Mar;15(3):141-56. doi: 10.1038/nrn3670. Nat Rev Neurosci. 2014. PMID: 24552784 Review.
References
- Arnold DB, Clapham DE. Molecular determinants for subcellular localization of PSD-95 with an interacting K+ channel. Neuron. 1999;23:149–157. - PubMed
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- OD 006117/OD/NIH HHS/United States
- R37 NS046579/NS/NINDS NIH HHS/United States
- NS-046579/NS/NINDS NIH HHS/United States
- GM 060416/GM/NIGMS NIH HHS/United States
- R01 GM060416/GM/NIGMS NIH HHS/United States
- R01 CA170820/CA/NCI NIH HHS/United States
- R01 AI085583/AI/NIAID NIH HHS/United States
- MH-086381/MH/NIMH NIH HHS/United States
- R01 NS046579/NS/NINDS NIH HHS/United States
- R01 NS069720/NS/NINDS NIH HHS/United States
- R01 GM053395/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- GM-083898/GM/NIGMS NIH HHS/United States
- GM53395/GM/NIGMS NIH HHS/United States
- R01 MH086381/MH/NIMH NIH HHS/United States
- R01 GM083898/GM/NIGMS NIH HHS/United States
- NS69720/NS/NINDS NIH HHS/United States
- R01 NS081678/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials