Prion-like propagation of protein aggregation and related therapeutic strategies - PubMed (original) (raw)
Review
Prion-like propagation of protein aggregation and related therapeutic strategies
Sarah K Kaufman et al. Neurotherapeutics. 2013 Jul.
Abstract
Many neurodegenerative diseases are characterized by the progressive accumulation of aggregated protein. Recent evidence suggests the prion-like propagation of protein misfolding underlies the spread of pathology observed in these diseases. This review traces our understanding of the mechanisms that underlie this phenomenon and discusses related therapeutic strategies that derive from it.
Figures
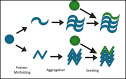
Fig. 1
Amplification of a protein aggregate by templated conformational change. A native protein (blue circle) adopts a pathological conformation that facilitates aggregation into b-sheet rich structures. These structures can contact additional native proteins (green circles), adding them on to the aggregate by converting them to a specific aggregate structure
Fig. 2
Mechanisms of aggregate release and uptake. a) Mechanisms of aggregate release. Proteins that misfold and aggregate might be secreted via several mechanisms. Direct penetration and vesicle fusion release aggregates directly into the media, whereas multivesicular body secretion and direct budding produce aggregate-laden vesicles. Therapeutic interventions can target the production of aggregate-prone proteins through RNA interference and antisense oligonucleotides. Small molecules might stabilize the native protein structure or inhibit templated misfolding of the cognate monomer. MVB, multivesicular body. b) Mechanisms of aggregate uptake. Fibrils are endocytosed by vesicle fusion or fluid-phase endocytosis. Aggregate escape into the cytosol allows further seeding and aggregation of endogenously expressed protein. Antibody blockade of cell attachment or endocytosis of aggregates will prevent the spread of misfolded protein, and therapies that induce relevant degradation pathways may increase the clearance of aggregates from the cell
Similar articles
- Prion-like mechanisms in neurodegenerative diseases.
Frost B, Diamond MI. Frost B, et al. Nat Rev Neurosci. 2010 Mar;11(3):155-9. doi: 10.1038/nrn2786. Epub 2009 Dec 23. Nat Rev Neurosci. 2010. PMID: 20029438 Free PMC article. Review. - Prion-like propagation of α-synuclein in neurodegenerative diseases.
Tarutani A, Hasegawa M. Tarutani A, et al. Prog Mol Biol Transl Sci. 2019;168:323-348. doi: 10.1016/bs.pmbts.2019.07.005. Epub 2019 Jul 31. Prog Mol Biol Transl Sci. 2019. PMID: 31699325 Review. - Neurodegeneration. Could they all be prion diseases?
Miller G. Miller G. Science. 2009 Dec 4;326(5958):1337-9. doi: 10.1126/science.326.5958.1337. Science. 2009. PMID: 19965731 No abstract available. - Prion-like mechanisms in the pathogenesis of tauopathies and synucleinopathies.
Goedert M, Falcon B, Clavaguera F, Tolnay M. Goedert M, et al. Curr Neurol Neurosci Rep. 2014 Nov;14(11):495. doi: 10.1007/s11910-014-0495-z. Curr Neurol Neurosci Rep. 2014. PMID: 25218483 Review. - [The Propagation Hypothesis of Prion-like Protein Agregates in Neurodegenerative Diseases].
Nonaka T. Nonaka T. Brain Nerve. 2019 Nov;71(11):1209-1214. doi: 10.11477/mf.1416201430. Brain Nerve. 2019. PMID: 31722306 Japanese.
Cited by
- Potential Pathways of Abnormal Tau and α-Synuclein Dissemination in Sporadic Alzheimer's and Parkinson's Diseases.
Braak H, Del Tredici K. Braak H, et al. Cold Spring Harb Perspect Biol. 2016 Nov 1;8(11):a023630. doi: 10.1101/cshperspect.a023630. Cold Spring Harb Perspect Biol. 2016. PMID: 27580631 Free PMC article. Review. - The olfactory bulb as the entry site for prion-like propagation in neurodegenerative diseases.
Rey NL, Wesson DW, Brundin P. Rey NL, et al. Neurobiol Dis. 2018 Jan;109(Pt B):226-248. doi: 10.1016/j.nbd.2016.12.013. Epub 2016 Dec 20. Neurobiol Dis. 2018. PMID: 28011307 Free PMC article. - Chronic exposure to cerebrospinal fluid of multiple system atrophy in neuroblastoma and glioblastoma cells induces cytotoxicity via ER stress and autophagy activation.
Wang X, Ma M, Teng J, Zhang J, Zhou S, Zhang Y, Wu E, Ding X. Wang X, et al. Oncotarget. 2015 May 30;6(15):13278-94. doi: 10.18632/oncotarget.3748. Oncotarget. 2015. PMID: 25965819 Free PMC article. - Key Points Concerning Amyloid Infectivity and Prion-Like Neuronal Invasion.
Espargaró A, Busquets MA, Estelrich J, Sabate R. Espargaró A, et al. Front Mol Neurosci. 2016 Apr 22;9:29. doi: 10.3389/fnmol.2016.00029. eCollection 2016. Front Mol Neurosci. 2016. PMID: 27147962 Free PMC article. - Human prion protein-induced autophagy flux governs neuron cell damage in primary neuron cells.
Moon JH, Lee JH, Nazim UM, Lee YJ, Seol JW, Eo SK, Lee JH, Park SY. Moon JH, et al. Oncotarget. 2016 May 24;7(21):29989-30002. doi: 10.18632/oncotarget.8802. Oncotarget. 2016. PMID: 27102156 Free PMC article.
References
- Caughey BW, Dong A, Bhat KS, Ernst D, Hayes SF, Caughey WS. Secondary structure analysis of the scrapie-associated protein PrP 27–30 in water by infrared spectroscopy. Biochemistry. 1991;30(31):7672–7680. - PubMed
- Büeler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, et al. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73(7):1339–1347. - PubMed
- Scott M, Foster D, Mirenda C, Serban D, Coufal F, Wälchli M, et al. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell. 1989;59(5):847–857. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical