Identification of an intact ParaHox cluster with temporal colinearity but altered spatial colinearity in the hemichordate Ptychodera flava - PubMed (original) (raw)
doi: 10.1186/1471-2148-13-129.
Yi-Chih Chen, Rossella Annunziata, Hsiu-Chi Ting, Che-huang Tung, Ryo Koyanagi, Kunifumi Tagawa, Tom Humphreys, Asao Fujiyama, Hidetoshi Saiga, Nori Satoh, Jr-Kai Yu, Maria Ina Arnone, Yi-Hsien Su
Affiliations
- PMID: 23802544
- PMCID: PMC3698058
- DOI: 10.1186/1471-2148-13-129
Identification of an intact ParaHox cluster with temporal colinearity but altered spatial colinearity in the hemichordate Ptychodera flava
Tetsuro Ikuta et al. BMC Evol Biol. 2013.
Abstract
Background: ParaHox and Hox genes are thought to have evolved from a common ancestral ProtoHox cluster or from tandem duplication prior to the divergence of cnidarians and bilaterians. Similar to Hox clusters, chordate ParaHox genes including Gsx, Xlox, and Cdx, are clustered and their expression exhibits temporal and spatial colinearity. In non-chordate animals, however, studies on the genomic organization of ParaHox genes are limited to only a few animal taxa. Hemichordates, such as the Enteropneust acorn worms, have been used to gain insights into the origins of chordate characters. In this study, we investigated the genomic organization and expression of ParaHox genes in the indirect developing hemichordate acorn worm Ptychodera flava.
Results: We found that P. flava contains an intact ParaHox cluster with a similar arrangement to that of chordates. The temporal expression order of the P. flava ParaHox genes is the same as that of the chordate ParaHox genes. During embryogenesis, the spatial expression pattern of PfCdx in the posterior endoderm represents a conserved feature similar to the expression of its orthologs in other animals. On the other hand, PfXlox and PfGsx show a novel expression pattern in the blastopore. Nevertheless, during metamorphosis, PfXlox and PfCdx are expressed in the endoderm in a spatially staggered pattern similar to the situation in chordates.
Conclusions: Our study shows that P. flava ParaHox genes, despite forming an intact cluster, exhibit temporal colinearity but lose spatial colinearity during embryogenesis. During metamorphosis, partial spatial colinearity is retained in the transforming larva. These results strongly suggest that intact ParaHox gene clustering was retained in the deuterostome ancestor and is correlated with temporal colinearity.
Figures
Figure 1
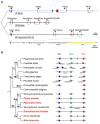
Genomic organizations of ParaHox genes. (A) Phylogenetic tree depicting genomic organization of the P. flava ParaHox cluster (top) compared to the clusters of amphioxus (Branchiostoma floridae) and mouse (Mus musculus) on chromosome (Chr.) 5. Blue, green, red, purple, and yellow boxes are exons of Gsx (Gsh), Xlox (Ipf1), Cdx, PRHOXNB, and Flt1, respectively. The light blue line indicates the fully sequenced PfBS11F10 BAC clone. Bent arrows indicate transcriptional orientations. The bottom line is a zoomed out view showing the neighboring Flt1 gene located near the mouse ParaHox cluster. (B) Evolution of the ParaHox cluster in bilateria. The phylogenetic tree represents the genomic organization of ParaHox genes in several bilateria, including protostomes (Pr) and deuterostomes (De). Protostomes can be further divided into lophotrochozoa (Lo) and ecdysozoa (Ed). The following lophotrochozoa animals were included in the analysis: two polychaete species (Platynereis dumerilii and Capitella teleta) and one leech species (Helobdella robusta) belonging to the Phylum Annelida (An); limpet (Lottia gigantea) and two oyster species (Crassostrea gigas and Pinctada fucata) in the Phylum Mollusca (Mo). Two ecdysozoa species fruit fly Drosophila melanogaster and nematode Caenorhabditis elegans do not contain the full complement of ParaHox genes. In deuterostomes, Phylum Echinodermata (Ec), including sea urchin Strongylocentrotus purpuratus and starfish Patiria miniata, and Phylum Hemichordata (He), including Ptychodera flava and Saccoglossus kowalevskii, constitute Ambulacraria (Am) that is closely related to Phylum Chordata (Ch). ParaHox gene organizations of the three chordate species, amphioxus Branchiostoma floridae, ascidian Ciona intestinalis, and mouse Mus musculus, are presented. IDs of the scaffolds (Sc) on which ParaHox genes are located are indicated beneath the illustrated scaffolds found in the genome databases. Blue, green, and red triangles indicate the orientations of Gsx, Xlox, and Cdx, respectively, in the genome. Double slashes between two genes indicate that although the genes are located on the same chromosome or scaffold they are separated by intervening genes. Species names shown in red contain intact ParaHox clusters.
Figure 2
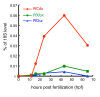
Temporal expression profiles of ParaHox genes. Transcript levels of ParaHox genes were measured by QPCR at different embryonic stages including unfertilized egg (0 hpf), early blastula (12 hpf), late blastula (16 hpf), early gastrula (24 hpf), late gastrula (43 hpf), and tornaria larva (65 hpf). The Y-axis is the transcript level normalized to the 18S rRNA level at the same stage.
Figure 3
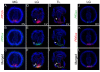
Expression patterns of P. flava ParaHox genes. In situ hybridization analyses of PfCdx (A-H), PfXlox (I-P), and PfGsx (Q-X) were performed on unfertilized egg (UF), blastula (B), early gastrula (EG), mid-gastrula (MG), late gastrula (LG), and tornaria larva (TL) at different time points (hpf, hours post fertilization; dpf, days post fertilization). White and black arrows indicate the expression of PfCdx and the midgut-hindgut boundary, respectively. Black and white arrowheads denote the expression of PfXlox and PfGsx, respectively. The observed views are indicated at the bottom right corner of the panels in the first row. Unless otherwise indicated, the observed views of subsequent panels are the same as the first panel in each column. DV, dorsal view; LV, lateral view; VV, ventral view. The scale bar is 50 μm. Panels T’ and U’ are the blastopore view (BV) of panels T and U, respectively.
Figure 4
Double fluorescent in situ hybridization of PfXlox with PfCdx or PfGsx . DNP-labeled PfXlox probe and DIG-labeled PfCdx probe were used for in situ hybridization on mid-gastrula (A-C), late gastrula (D-F), and tornaria larva (G-I). The arrows and arrowheads indicate the anterior boundaries of the PfCdx and PfXlox expression domain, respectively. Double stainings of PfGsx and PfXlox transcripts were performed on the late gastrula embryos (J-L).
Figure 5
Expression of PfCdx and PfXlox in transforming larvae. Whole mount (A, D) and sagittal sections (B, E) of the transforming larvae hybridized with antisense probes against PfCdx (A-C) or PfXlox (D-F). Panels C and F are higher magnifications of panel B and E, respectively. Arrows and arrowheads indicate the in situ hybridization signals of PfCdx and PfXlox, respectively. The scale bar is 200 μm.
Similar articles
- Intact cluster and chordate-like expression of ParaHox genes in a sea star.
Annunziata R, Martinez P, Arnone MI. Annunziata R, et al. BMC Biol. 2013 Jun 27;11:68. doi: 10.1186/1741-7007-11-68. BMC Biol. 2013. PMID: 23803323 Free PMC article. - Time is of the essence for ParaHox homeobox gene clustering.
Garstang M, Ferrier DE. Garstang M, et al. BMC Biol. 2013 Jun 26;11:72. doi: 10.1186/1741-7007-11-72. BMC Biol. 2013. PMID: 23803337 Free PMC article. Review. - Genetic organization and embryonic expression of the ParaHox genes in the sea urchin S. purpuratus: insights into the relationship between clustering and colinearity.
Arnone MI, Rizzo F, Annunciata R, Cameron RA, Peterson KJ, Martínez P. Arnone MI, et al. Dev Biol. 2006 Dec 1;300(1):63-73. doi: 10.1016/j.ydbio.2006.07.037. Epub 2006 Aug 4. Dev Biol. 2006. PMID: 16959236 - Identical genomic organization of two hemichordate hox clusters.
Freeman R, Ikuta T, Wu M, Koyanagi R, Kawashima T, Tagawa K, Humphreys T, Fang GC, Fujiyama A, Saiga H, Lowe C, Worley K, Jenkins J, Schmutz J, Kirschner M, Rokhsar D, Satoh N, Gerhart J. Freeman R, et al. Curr Biol. 2012 Nov 6;22(21):2053-8. doi: 10.1016/j.cub.2012.08.052. Epub 2012 Oct 11. Curr Biol. 2012. PMID: 23063438 Free PMC article. - Evolution of invertebrate deuterostomes and Hox/ParaHox genes.
Ikuta T. Ikuta T. Genomics Proteomics Bioinformatics. 2011 Jun;9(3):77-96. doi: 10.1016/S1672-0229(11)60011-9. Genomics Proteomics Bioinformatics. 2011. PMID: 21802045 Free PMC article. Review.
Cited by
- The TALE face of Hox proteins in animal evolution.
Merabet S, Galliot B. Merabet S, et al. Front Genet. 2015 Aug 18;6:267. doi: 10.3389/fgene.2015.00267. eCollection 2015. Front Genet. 2015. PMID: 26347770 Free PMC article. - Intact cluster and chordate-like expression of ParaHox genes in a sea star.
Annunziata R, Martinez P, Arnone MI. Annunziata R, et al. BMC Biol. 2013 Jun 27;11:68. doi: 10.1186/1741-7007-11-68. BMC Biol. 2013. PMID: 23803323 Free PMC article. - BMP controls dorsoventral and neural patterning in indirect-developing hemichordates providing insight into a possible origin of chordates.
Su YH, Chen YC, Ting HC, Fan TP, Lin CY, Wang KT, Yu JK. Su YH, et al. Proc Natl Acad Sci U S A. 2019 Jun 25;116(26):12925-12932. doi: 10.1073/pnas.1901919116. Epub 2019 Jun 12. Proc Natl Acad Sci U S A. 2019. PMID: 31189599 Free PMC article. - TCF/Lef regulates the Gsx ParaHox gene in central nervous system development in chordates.
Garstang MG, Osborne PW, Ferrier DE. Garstang MG, et al. BMC Evol Biol. 2016 Mar 3;16:57. doi: 10.1186/s12862-016-0614-3. BMC Evol Biol. 2016. PMID: 26940763 Free PMC article. - Time is of the essence for ParaHox homeobox gene clustering.
Garstang M, Ferrier DE. Garstang M, et al. BMC Biol. 2013 Jun 26;11:72. doi: 10.1186/1741-7007-11-72. BMC Biol. 2013. PMID: 23803337 Free PMC article. Review.
References
- Ferrier DE, Holland PW. Ancient origin of the Hox gene cluster. Nat Rev Genet. 2001;2:33–38. - PubMed
- Garcia-Fernandez J. The genesis and evolution of homeobox gene clusters. Nat Rev Genet. 2005;6:881–892. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources