Sustained production of ROS triggers compensatory proliferation and is required for regeneration to proceed - PubMed (original) (raw)
Sustained production of ROS triggers compensatory proliferation and is required for regeneration to proceed
Carole Gauron et al. Sci Rep. 2013.
Abstract
A major issue in regenerative medicine is the role of injury in promoting cell plasticity. Here we explore the function of reactive oxygen species (ROS) induced through lesions in adult zebrafish. We show that ROS production, following adult fin amputation, is tightly regulated in time and space for at least 24 hours, whereas ROS production remains transient (2 hours) in mere wound healing. In regenerative tissue, ROS signaling triggers two distinct parallel pathways: one pathway is responsible for apoptosis, and the other pathway is responsible for JNK activation. Both events are involved in the compensatory proliferation of stump epidermal cells and are necessary for the progression of regeneration. Both events impact the Wnt, SDF1 and IGF pathways, while apoptosis only impacts progenitor marker expression. These results implicate oxidative stress in regeneration and provide new insights into the differences between healing and regeneration.
Figures
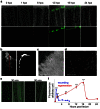
Figure 1. Sustained production of ROS is specific to regeneration.
(a–e) ROS were detected with a fluorescent probe (H2DCFDA) in the time course of regeneration (a–d) or during wounding (e). (a) Top row: merge; bottom row: ROS detection. (b) At 16 hpa and higher magnification, orthogonal projection showed that ROS were primarily detected in the epidermis. (c–d) At 16 hpa, ROS were abundant in the wounded epidermis (c) and decorated adjacent epidermal cells (d). (e) Left panel: ROS detection; right panel: merge. (f) Quantification of the amount of ROS during healing and regeneration. Error bars represent the SEM (**p< 0.01, ***p < 0.001). Scale bars = 20 μm.
Figure 2. Sustained ROS production is essential for regeneration to proceed.
(a) Inhibition of NADPH oxidase with VAS2870 1 μM or DPI 1 μM reduced the size of the regenerate at 72 hpa. Representative images are shown. The efficiency of regeneration was quantified at 3 dpa (day post amputation). The surface of the blastema was measured and subsequently divided using the square length of the amputation plane for each fish. The efficiency of regeneration is expressed as a percentage of the control. (b) Gene expression was analyzed through quantitative RT-PCR on the regenerated fin at 18 hpa after VAS2870 (1 μM) or vehicle (DMSO) treatment. The DMSO-treated sample was taken as 1. (c, d) Quantification of phospho-c-JUN was performed on western blots. Full-length western blots are presented in Supplementary Figure 5. (d) The inhibition of NADPH oxidases (NOX) with VAS2870 (1 μM) or JNK activity with SP600125 (5 μM) reduced c-Jun phosphorylation at 6 hpa. (e) The inhibition of ROS (VAS2870 1 μM) production decreased apoptosis at 18 hpa. In representative pictures, dotted lines indicate amputation plane. The error bars represent the SEM (* p<0.05, **p< 0.01, ***p < 0.001).
Figure 3. Spatio-temporal control of cell death during regeneration and wounding (a–c) TUNEL staining during the time course of regeneration.
For each time point, a higher magnification is shown for the distal (segment 1 and 2, blue frame) and proximal (segment 3–4, red frame) regions of the fin. Numerous TUNEL-positive cells were detected in the proximal region of the fin (segments 3–6) at 18 hpa (arrowheads). (b) Quantification of cell death. For each fin, TUNEL-positive cells (gray) or active Caspase-3 positive cells (black) were counted in ray and inter-ray 2 in all segments. The uncut control (nc) corresponds to positive cells below the level of amputation in amputated fins (first bifurcation level) and was taken as 1. (c) Longitudinal cross-sections obtained at 18 hpa revealed numerous positive cells in the stump epidermis (arrows) and only a few cells in the mesenchyme (arrowhead). (d, e) Time course of cell death during wounding. TUNEL staining was performed over the indicated time period. The error bars represent the SEM (* p<0.05, **p< 0.01, ***p < 0.001).
Figure 4. Inhibition of apoptosis impairs blastema formation.
(a, b) TUNEL assay and immuno-detection of active Caspase-3 were performed on 18-hpa samples (white: DMSO; grey: NS3694 5 μM). (c) The efficiency of regeneration was quantified at 72 hpa and is expressed as a percentage of control; representative fins are shown. (d) Fish were incubated in M50054 (a cell permeable inhibitor of apoptosis) at 30 μM or vehicle (DMSO) from the time of amputation to 3 dpa. The efficiency of regeneration was quantified at 3 dpa and is expressed as a percentage of the control. (e) The inhibition of the first wave of apoptosis does not affect the second wave. Fish were incubated in NS3694 (5 μM) or vehicle (DMSO) from 4 hours before amputation to 12 hpa. The fish were subsequently transferred to water until 18 hpa. The fins were collected, and the immuno-detection of active Caspase-3 was performed. Positives cells were counted in ray and inter ray 2 for each fin. No significant differences were observed. (f) Hematoxylin-stained longitudinal sections of the regenerated fin at 3 dpa with or without NS3694 treatment. In representative pictures, the dotted lines indicate amputation plane. The error bars represent the SEM (* p<0.05, **p< 0.01, ***p < 0.001).
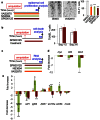
Figure 5. JNK and apoptosis are involved in compensatory proliferation.
(a) Inhibition of ROS production (VAS2870 1 μM), Apoptosis (NS3496 5 μM) and JNK (SP600125 5 μM) reduced the number of epidermal cells in mitosis at 24 hpa. Proliferation in the epidermis was monitored through the phosphorylated histone H3 (H3-P). Representative pictures are shown. H3-P positive cells were counted in rays and inter-rays-2 from segment 1 to segment 6 at 24 hpa. (b) Inhibition of JNK (SP600125 5 μM) had no effect on cell death at 18 hpa. (c–e) Gene expression was analyzed through quantitative RT-PCR on the regenerated fin at 18 hpa after NS3694 (5 μM), (SP600125 5 μM), VAS2870 (1 μM) or vehicle (DMSO) treatment. The DMSO-treated sample was taken as 1. The error bars represent the SEM (* p<0.05, **p< 0.01, ***p < 0.001).
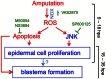
Figure 6. Schematic representation of early events following amputation.
Amputation triggers a sustained ROS production that induces apoptosis and JNK signaling. Both signaling pathways are important for epidermal cell proliferation at 24 hpa and are necessary for blastema formation. ROS through Apoptosis signaling stimulates pluripotency marker expression.
Similar articles
- Cell death: a program to regenerate.
Vriz S, Reiter S, Galliot B. Vriz S, et al. Curr Top Dev Biol. 2014;108:121-51. doi: 10.1016/B978-0-12-391498-9.00002-4. Curr Top Dev Biol. 2014. PMID: 24512708 Review. - Toxicity of silver nanoparticles on wound healing: A case study of zebrafish fin regeneration model.
Pang S, Gao Y, Wang F, Wang Y, Cao M, Zhang W, Liang Y, Song M, Jiang G. Pang S, et al. Sci Total Environ. 2020 May 15;717:137178. doi: 10.1016/j.scitotenv.2020.137178. Epub 2020 Feb 7. Sci Total Environ. 2020. PMID: 32062274 - Retinoic acid signaling controls the formation, proliferation and survival of the blastema during adult zebrafish fin regeneration.
Blum N, Begemann G. Blum N, et al. Development. 2012 Jan;139(1):107-16. doi: 10.1242/dev.065391. Epub 2011 Nov 17. Development. 2012. PMID: 22096078 - Mechanistic target of rapamycin complex 1 signaling regulates cell proliferation, cell survival, and differentiation in regenerating zebrafish fins.
Hirose K, Shiomi T, Hozumi S, Kikuchi Y. Hirose K, et al. BMC Dev Biol. 2014 Dec 6;14:42. doi: 10.1186/s12861-014-0042-9. BMC Dev Biol. 2014. PMID: 25480380 Free PMC article. - An unexpected friend - ROS in apoptosis-induced compensatory proliferation: Implications for regeneration and cancer.
Diwanji N, Bergmann A. Diwanji N, et al. Semin Cell Dev Biol. 2018 Aug;80:74-82. doi: 10.1016/j.semcdb.2017.07.004. Epub 2017 Jul 5. Semin Cell Dev Biol. 2018. PMID: 28688927 Free PMC article. Review.
Cited by
- Finding Solutions for Fibrosis: Understanding the Innate Mechanisms Used by Super-Regenerator Vertebrates to Combat Scarring.
Durant F, Whited JL. Durant F, et al. Adv Sci (Weinh). 2021 Aug;8(15):e2100407. doi: 10.1002/advs.202100407. Epub 2021 May 24. Adv Sci (Weinh). 2021. PMID: 34032013 Free PMC article. Review. - Age-dependent decline in fin regenerative capacity in the short-lived fish Nothobranchius furzeri.
Wendler S, Hartmann N, Hoppe B, Englert C. Wendler S, et al. Aging Cell. 2015 Oct;14(5):857-66. doi: 10.1111/acel.12367. Epub 2015 Jun 29. Aging Cell. 2015. PMID: 26121607 Free PMC article. - Mitochondrial DNA Repair in Neurodegenerative Diseases and Ageing.
Bazzani V, Equisoain Redin M, McHale J, Perrone L, Vascotto C. Bazzani V, et al. Int J Mol Sci. 2022 Sep 27;23(19):11391. doi: 10.3390/ijms231911391. Int J Mol Sci. 2022. PMID: 36232693 Free PMC article. Review. - A Spatiotemporal Characterisation of Redox Molecules in Planarians, with a Focus on the Role of Glutathione during Regeneration.
Bijnens K, Jaenen V, Wouters A, Leynen N, Pirotte N, Artois T, Smeets K. Bijnens K, et al. Biomolecules. 2021 May 11;11(5):714. doi: 10.3390/biom11050714. Biomolecules. 2021. PMID: 34064618 Free PMC article. - Drosophila Imaginal Discs as a Model of Epithelial Wound Repair and Regeneration.
Smith-Bolton R. Smith-Bolton R. Adv Wound Care (New Rochelle). 2016 Jun 1;5(6):251-261. doi: 10.1089/wound.2014.0547. Adv Wound Care (New Rochelle). 2016. PMID: 27274435 Free PMC article. Review.
References
- Brockes J. P. & Kumar A. Comparative aspects of animal regeneration. Annu Rev Cell Dev Biol 24, 525–549 (2008). - PubMed
- Galliot B. & Ghila L. Cell plasticity in homeostasis and regeneration. Mol Reprod Dev 77, 837–855 (2010). - PubMed
- Nakatani Y., Kawakami A. & Kudo A. Cellular and molecular processes of regeneration, with special emphasis on fish fins. Dev Growth Differ 49, 145–154 (2007). - PubMed
- Brockes J. P. Amphibian limb regeneration: rebuilding a complex structure. Science 276, 81–87 (1997). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials