OTULIN restricts Met1-linked ubiquitination to control innate immune signaling - PubMed (original) (raw)
OTULIN restricts Met1-linked ubiquitination to control innate immune signaling
Berthe Katrine Fiil et al. Mol Cell. 2013.
Abstract
Conjugation of Met1-linked polyubiquitin (Met1-Ub) by the linear ubiquitin chain assembly complex (LUBAC) is an important regulatory modification in innate immune signaling. So far, only few Met1-Ub substrates have been described, and the regulatory mechanisms have remained elusive. We recently identified that the ovarian tumor (OTU) family deubiquitinase OTULIN specifically disassembles Met1-Ub. Here, we report that OTULIN is critical for limiting Met1-Ub accumulation after nucleotide-oligomerization domain-containing protein 2 (NOD2) stimulation, and that OTULIN depletion augments signaling downstream of NOD2. Affinity purification of Met1-Ub followed by quantitative proteomics uncovered RIPK2 as the predominant NOD2-regulated substrate. Accordingly, Met1-Ub on RIPK2 was largely inhibited by overexpressing OTULIN and was increased by OTULIN depletion. Intriguingly, OTULIN-depleted cells spontaneously accumulated Met1-Ub on LUBAC components, and NOD2 or TNFR1 stimulation led to extensive Met1-Ub accumulation on receptor complex components. We propose that OTULIN restricts Met1-Ub formation after immune receptor stimulation to prevent unwarranted proinflammatory signaling.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
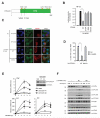
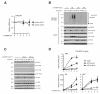
Figure 1. OTULIN regulates signaling in response to NOD2 stimulation
(A) Schematic depiction of human OTULIN. Circles illustrate residues required for ubiquitin binding and stars denote residues required for cleavage of Met1-Ub. (B) NF-κB activity in HEK293T cell lysates transfected with HA-NOD2 alone or with OTULINWT or the indicated point mutants. (C) Immunofluorescence analysis of nuclear translocation of the NF-κB subunit RelA/p65 (green) in response to L18-MDP stimulation (1 μg/mL) in U2OS/NOD2 cells transfected with empty vector (pcDNA3-3xHA; HA-vector), OTULINWT or OTULINWA/CA. Scalebar, 10 μm (D) Quantification of nuclear NF-κB translocation after L18-MDP stimulation of cells treated as in (C). (E) Relative levels of TNF, IL8 and IL6 transcripts measured by qRT-PCR on cDNA from U2OS/NOD2 control and OTULIN-depleted cells treated with L18-MDP (200 ng/mL). Immunoblot of OTULIN levels in control (MM) and siOTULIN-treated cells. (F) Immunoblotting of IκBα degradation and phosphorylation of signaling components in response to stimulation with L18-MDP (200 ng/mL) in control and OTULIN-depleted cells. Data in (B, D and E) represent the mean ± SEM of at least three independent experiments, each performed in duplicate. In (D) at least 150 cells were counted per condition in each experiment. Asterisks (**) indicates p < 0.01. See also Figure S1.
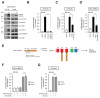
Figure 2. OTULIN is part of the NOD2 receptor complex and antagonizes Met1-Ub-dependent signaling
(A) Immunoprecipitation with anti-HA in U2OS/NOD2 cells. HA-NOD2 expression was induced for 24 hours with doxycycline (DOX). Immunoprecipitates were examined for co-purification of OTULIN and known members of the NOD2 receptor complex. (B-D) NF-κB activity in HEK293T cell lysates transfected with luciferase reporters and OTULIN, LUBAC (HOIP, HOIL-1), DN-LUBAC (RING-mutated HOIP, HOIL-1) or XIAP as indicated. (E) Schematic depiction of the engineered AVPI-Ub4GS protein, which binds to XIAP BIR2 and BIR3 domains via an N-terminal IBM to activate NF-κB. DUB, deubiquitinase. (F, G) NF-κB activity in HEK293T cell lysates transfected luciferase reporters and XIAPF/A, AVPI-Ub4GS, DN-LUBAC or OTULIN as indicated. Data in (B-D and F-G) represent the mean ± SEM of at least three independent experiments (except ‘OTULIN’ in 2B where n = 2), each performed in duplicate. Asterisks (**) indicates p < 0.01. n.s., not significant. See also Figure S2.
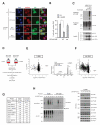
Figure 3. Proteome-wide quantification of NOD2-regulated Met1-Ub
(A) Immunofluorescence analysis of nuclear translocation of the NF-κB subunit RelA/p65 (red) in response to L18-MDP stimulation (1 μg/mL) in U2OS/NOD2 cells transfected with GFP vector, GFP-M1-SUB, or a Ub-binding mutant GFP-M1-SUBmut. Scalebar, 10 μm (B) Quantification of nuclear NF-κB translocation after L18-MDP stimulation of cells treated as in (A). (C) Purification of endogenous Ub conjugates using GST-coupled M1-SUB (Met1-Ub), or TUBE (all Ub-linkages). Pulldown with glutathione sepharose beads on HEK293T cell lysates transfected with LUBAC (HOIP and HOIL) and the indicated OTULIN mutants. Purified material and lysate was examined by immunoblotting. (D) Outline of the SILAC-based proteomics approach for quantification of protein ubiquitination in response to NOD2 stimulation. (E) MS data from TUBE1-purification of ubiquitinated proteins from THP-1 cells treated or not with L18-MDP (200 ng/mL) for 60 min. The circle representing RIPK2 is marked in red. The mass spectrum shows the relative abundance of the RIPK2 peptide TQNILLDNEFHVK in L18-MDP treated (SILAC heavy) cells compared to untreated (SILAC light). (F) Same as in (E) but after purification of ubiquitinated proteins with M1-SUB. RIPK2 is marked in red. (G) SILAC H/L ratios of proteins identified in (E) and (F) with reported function in the NOD2 signaling complex. NA, not available. (H) Time course analysis of RIPK2 ubiquitination with TUBE and M1-SUB in lysates of U2OS/NOD2 cells treated with L18-MDP (200 ng/mL). Purified material and lysate was examined by immunoblotting. Data in (B) represent the mean ± SEM of three independent experiments. At least 140 cells were counted per condition in each experiment. Asterisks (*) indicates p < 0.05. See also Figure S3 and Table S1.
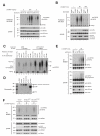
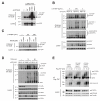
Figure 4. OTULIN limits Met1-linked ubiquitination of RIPK2 after NOD2 stimulation
(A, B) Purification of endogenous Ub conjugates with M1-SUB from (A) OTULIN-depleted U2OS/NOD2 cells treated with either 200 ng/mL L18-MDP for 1 hour or 5 ng/mL L18-MDP for 2 hours (B) OTULIN overexpressing U2OS/NOD2 cells treated with L18-MDP (200 ng/mL) for the indicated times. Purified material and lysate from (A, B) was examined by immunoblotting. (C) Ub-chain restriction analysis of ubiquitinated RIPK2 isolated with M1-SUB from L18-MDP treated and siMM or siOTULIN transfected cells. Purified Ub-conjugates were incubated with the indicated deubiquitinases (DUBs) for 1h, and samples were examined by immunoblotting. (D) Coomassie staining of the recombinant DUBs used in (C). (E) Immunoprecipitation of endogenous NEMO from OTULIN-depleted cells treated with 200 ng/mL L18-MDP for 1 hour. Purified material and lysate was examined by immunoblotting (F) Immunoprecipitation of endogenous NEMO from OTULIN overexpressing cells treated with 200 ng/mL L18-MDP for 1 hour or 5 ng/mL L18-MDP for 2 hours. Purified material and lysate was examined by immunoblotting.
Figure 5. OTULIN regulates early NOD2 signaling and sensitivity to L18-MDP
(A) Relative levels of OTULIN transcripts measured by qRT-PCR with two different primer sets on cDNA from U2OS/NOD2 control cells treated with 200 ng/mL L18-MDP for the times indicated. (B) Purification of endogenous Ub conjugates with M1-SUB from OTULIN-depleted cells treated with L18-MDP for up to 4 h. Purified material and lysate was examined by immunoblotting. (C) Immunoblotting of IκBα degradation and phosphorylation of signaling components in response to stimulation with 5 ng/mL of L18-MDP in control and OTULIN-depleted U2OS/NOD2 cells. (D) Relative levels of TNF, IL8 and IL6 transcripts measured by qRT-PCR on cDNA from U2OS/NOD2 control and OTULIN-depleted cells treated with 5 ng/mL of L18-MDP. Data in (A, D) represent the mean ± SEM of at least three independent experiments. See also Figure S4.
Figure 6. Decreased OTULIN function leads to promiscuous Met1-Ub accumulation
(A-C) Samples used in Figure 3A (C), 4A (B), and 4B (C) were immunoblotted for LUBAC components as indicated. (D) Purification of endogenous Ub conjugates with M1-SUB in U2OS/NOD2 control and OTULIN-depleted cells treated with TNF (10 ng/mL). Purified material and lysate was examined by immunoblotting. (E) TNF-RSC purification by FLAG-TNF (100 ng/mL) immunoprecipitation from TREx293 control or OTULIN-depleted cells at different times after FLAG-TNF stimulation. Purified material was analyzed by immunoblotting. See also Figure S5.
Similar articles
- CYLD Limits Lys63- and Met1-Linked Ubiquitin at Receptor Complexes to Regulate Innate Immune Signaling.
Hrdinka M, Fiil BK, Zucca M, Leske D, Bagola K, Yabal M, Elliott PR, Damgaard RB, Komander D, Jost PJ, Gyrd-Hansen M. Hrdinka M, et al. Cell Rep. 2016 Mar 29;14(12):2846-58. doi: 10.1016/j.celrep.2016.02.062. Epub 2016 Mar 17. Cell Rep. 2016. PMID: 26997266 Free PMC article. - Met1-linked ubiquitination in immune signalling.
Fiil BK, Gyrd-Hansen M. Fiil BK, et al. FEBS J. 2014 Oct;281(19):4337-50. doi: 10.1111/febs.12944. Epub 2014 Aug 12. FEBS J. 2014. PMID: 25060092 Free PMC article. Review. - OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin.
Keusekotten K, Elliott PR, Glockner L, Fiil BK, Damgaard RB, Kulathu Y, Wauer T, Hospenthal MK, Gyrd-Hansen M, Krappmann D, Hofmann K, Komander D. Keusekotten K, et al. Cell. 2013 Jun 6;153(6):1312-26. doi: 10.1016/j.cell.2013.05.014. Cell. 2013. PMID: 23746843 Free PMC article. - Ubiquitin-dependent and -independent functions of OTULIN in cell fate control and beyond.
Weinelt N, van Wijk SJL. Weinelt N, et al. Cell Death Differ. 2021 Feb;28(2):493-504. doi: 10.1038/s41418-020-00675-x. Epub 2020 Dec 7. Cell Death Differ. 2021. PMID: 33288901 Free PMC article. Review. - LUBAC and OTULIN regulate autophagy initiation and maturation by mediating the linear ubiquitination and the stabilization of ATG13.
Chu Y, Kang Y, Yan C, Yang C, Zhang T, Huo H, Liu Y. Chu Y, et al. Autophagy. 2021 Jul;17(7):1684-1699. doi: 10.1080/15548627.2020.1781393. Epub 2020 Jun 26. Autophagy. 2021. PMID: 32543267 Free PMC article.
Cited by
- Lys63/Met1-hybrid ubiquitin chains are commonly formed during the activation of innate immune signalling.
Emmerich CH, Bakshi S, Kelsall IR, Ortiz-Guerrero J, Shpiro N, Cohen P. Emmerich CH, et al. Biochem Biophys Res Commun. 2016 Jun 3;474(3):452-461. doi: 10.1016/j.bbrc.2016.04.141. Epub 2016 Apr 29. Biochem Biophys Res Commun. 2016. PMID: 27133719 Free PMC article. - Deubiquitinases: Novel Therapeutic Targets in Immune Surveillance?
Lopez-Castejon G, Edelmann MJ. Lopez-Castejon G, et al. Mediators Inflamm. 2016;2016:3481371. doi: 10.1155/2016/3481371. Epub 2016 Aug 14. Mediators Inflamm. 2016. PMID: 27597804 Free PMC article. Review. - A bio-orthogonal linear ubiquitin probe identifies STAT3 as a direct substrate of OTULIN in glioblastoma.
Du X, Pang J, Gu B, Si T, Chang Y, Li T, Wu M, Wang Z, Wang Y, Feng J, Wu N, Man J, Li H, Li A, Zhang T, Wang B, Duan X. Du X, et al. Nucleic Acids Res. 2023 Feb 22;51(3):1050-1066. doi: 10.1093/nar/gkad002. Nucleic Acids Res. 2023. PMID: 36660824 Free PMC article. - Reciprocal interplay between OTULIN-LUBAC determines genotoxic and inflammatory NF-κB signal responses.
Li M, Li L, Asemota S, Kakhniashvili D, Narayanan R, Wang X, Liao FF. Li M, et al. Proc Natl Acad Sci U S A. 2022 Aug 16;119(33):e2123097119. doi: 10.1073/pnas.2123097119. Epub 2022 Aug 8. Proc Natl Acad Sci U S A. 2022. PMID: 35939695 Free PMC article. - The role of Ubiquitination in Apoptosis and Necroptosis.
Roberts JZ, Crawford N, Longley DB. Roberts JZ, et al. Cell Death Differ. 2022 Feb;29(2):272-284. doi: 10.1038/s41418-021-00922-9. Epub 2021 Dec 15. Cell Death Differ. 2022. PMID: 34912054 Free PMC article. Review.
References
- Bertrand MJ, Doiron K, Labbe K, Korneluk RG, Barker PA, Saleh M. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity. 2009;30:789–801. - PubMed
- Beug ST, Cheung HH, LaCasse EC, Korneluk RG. Modulation of immune signalling by inhibitors of apoptosis. Trends in immunology. 2012;33:535–545. - PubMed
- Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases