Hormone-regulated transcriptomes: lessons learned from estrogen signaling pathways in breast cancer cells - PubMed (original) (raw)
Review
Hormone-regulated transcriptomes: lessons learned from estrogen signaling pathways in breast cancer cells
Nasun Hah et al. Mol Cell Endocrinol. 2014.
Abstract
Recent rapid advances in next generation sequencing technologies have expanded our understanding of steroid hormone signaling to a genome-wide level. In this review, we discuss the use of a novel genomic approach, global nuclear run-on coupled with massively parallel sequencing (GRO-seq), to explore new facets of the steroid hormone-regulated transcriptome, especially estrogen responses in breast cancer cells. GRO-seq is a high throughput sequencing method adapted from conventional nuclear run-on methodologies, which is used to obtain a map of the position and orientation of all transcriptionally engaged RNA polymerases across the genome with extremely high spatial resolution. GRO-seq, which is an excellent tool for examining transcriptional responses to extracellular stimuli, has been used to comprehensively assay the effects of estrogen signaling on the transcriptome of ERα-positive MCF-7 human breast cancer cells. These studies have revealed new details about estrogen-dependent transcriptional regulation, including effects on transcription by all three RNA polymerases, complex transcriptional dynamics in response to estrogen signaling, and identification novel, unannotated non-coding RNAs. Collectively, these studies have been useful in discerning the molecular logic of the estrogen-regulated mitogenic response.
Keywords: 17β-estradiol; Breast cancer cells; ChIP; E2; ERBS; ERα; Estrogen; Estrogen receptor; GRO-seq; HMM; Non-coding RNA; Pol (I, II, III); RNA polymerase (I, II, III); SERM; Transcriptome; chromatin immunoprecipitation; eRNA; enhancer RNA; estrogen receptor alpha; estrogen receptor α binding site; global nuclear run-on and sequencing; hidden Markov model lncRNA, long non-coding RNAs; mRNA; messenger RNA; rRNA; ribosomal RNA; selective estrogen-receptor modulator; tRNA; transfer RNA.
Copyright © 2013 Elsevier Ireland Ltd. All rights reserved.
Figures
Figure 1. Assembly and function of enhancer complexes at estrogen receptor binding sites
Estrogen receptors (ERs) can bind to genomic DNA directly through estrogen response elements (EREs; green boxes) (top left) or indirectly by tethering through other transcription factors (e.g., AP-1, SP1, NF-κB) (bottom left). The binding of ER may be facilitated by the prior binding of pioneer factors, such as FOXA1 and AP2γ (top left). Upon binding to chromatin in response to estrogen, ER recruits co-regulators (e.g., Mediator, SRC, p300/CBP) to enhancers to promote transcription of target gene expression through short-range interactions (promoter proximal enhancers; − looping) or long-range interactions (promoter distal enhancers; + DNA looping) with target gene promoters (right). The outcome is the regulation of RNA polymerase II (Pol II) recruitment or activity, leading to increased target gene expression.
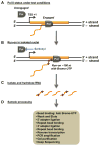
Figure 2. Protocol for GRO-seq
Global run-on coupled with massively parallel sequencing (GRO-seq) is a direct, high throughput sequencing method used to obtain a map of the position and orientation of all transcriptionally engaged RNA polymerases (Pols I, II, and III) across the genome [51]. (A) RNAs are transcribed throughout the genome by actively engaged RNA polymerases (Pol) in cells. (B) In order to map the location and density of RNA polymerases using GRO-seq, nuclei isolated from cells are subjected to a short run-on reaction in the presence of an NTP analog (Bromo-UTP) to label nascent RNAs. The reaction is performed in the presence of Sarkosyl, which blocks unengaged RNA polymerases from initiating transcription and removes impediments to elongating RNA polymerases during the run-on. Only newly synthesized portions of the RNA molecules are labeled with Bromo-UTP. (C) RNAs are isolated from the run-on reactions and base hydrolyzed to generate short fragments for short-read sequencing. (D) Bromo-UTP incorporated, newly synthesized RNAs are enriched by Bromo-UTP specific antibody conjugated-beads. Isolated nascent RNAs are ligated with RNA-adaptors, reverse-transcribed, and amplified for high throughput sequencing.
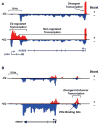
Figure 3. Genome browser views of GRO-seq data
Genome browser views of GRO-seq reads mapped to the human reference genome. The samples were prepared from ERα-positive MCF-7 human breast cancer cells treated with or without estradiol (E2) for 40 min [39,52]. Red indicates plus (+) strand reads and blue indicates minus (−) strand reads. Gene annotations with introns and exons are shown as schematics below the browser tracks. The transcription start sites (TSSs; bent arrows) and direction of transcription (repeated arrowheads) are shown. (A) Transcription of a protein coding gene. Upon stimulation with E2, transcription of LHX4 is induced, whereas transcription of ACBD6 is unregulated. The location of divergent promoter transcription is shown. (B) Transcription of the distal enhancer of the SMAD7 gene. Upon stimulation with E2, transcription of the SMAD7 distal ERα enhancer is up-regulated, generating enhancer RNAs (eRNAs). Transcription of the distal enhancer corresponds to transcription of the SMAD7 gene.
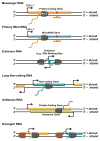
Figure 4. GRO-seq detects different types of Pol II-transcribed RNAs
A two state hidden Markov model (HMM)-based algorithm can be used with GRO-seq data to map transcription units that produce different types of RNAs. Each schematic illustrates a different type of transcription unit being transcribed by RNA polymerase II (Pol II). The transcription start sites (TSS; bent arrows), plus and minus DNA strands, actively transcribed RNAs, and orientation of the RNA polymerases are shown. Messenger RNAs (mRNAs) are 5′ capped, spliced, and polyadenylated protein coding RNAs, which may contain microRNAs embedded in their introns. MicroRNAs may also be transcribed as primary transcripts from an intergenic region using their own promoter. Enhancer RNAs (eRNAs) are short, typically divergently/bidirectionally transcribed RNAs that originate from enhancers, including ERα binding sites. Long non-coding RNAs (lncRNAs) are mRNA-like transcripts that are longer than 200 nucleotides, but lack coding potential. They are typically transcribed from intergenic regions, but their transcription units may overlap protein-coding genes. Antisense RNAs (asRNAs) originate from the opposite strand of an annotated protein coding or lncRNA gene with significant overlap of the transcribed regions. Lastly, divergent RNAs are short non-coding RNAs that are transcribed from thee opposite strand of a RefSeq gene promoter, other Pol II promoter, or enhancer. Non-coding RNAs transcribed by RNA Pol I and III are not show, but they are detected by GRO-seq.
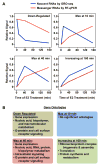
Figure 5. Dynamics and biology of an estrogen-dependent transcriptional response
GRO-seq analysis of an estrogen-dependent transcriptional response in ERα-positive MCF-7 human breast cancer cells subjected to a time course of estrogen treatment. (A) Estrogen- dependent transcriptional responses for protein coding genes can be classified into four distinct classes based on the dynamics of the response over a time course of estradiol (E2) treatment. The graphs are based on data from Hah et al. (2011) [52], but are stylized and are not factually quantitative. GRO-seq detects instantaneous changes in transcription that reflect the rapid actions of estrogen signaling on the transcriptome. Steady-state mRNA levels parallel the transcriptional response, but are clearly temporally delayed. (B) Gene ontology analyses of each class of genes from (A) reflect distinct biological effects the estrogen-dependent transcriptional response [52].
Figure 6. Determination of Pol II transcription rates and dynamics using GRO-seq
GRO-seq allows the monitoring of instantaneous changes in RNA polymerase location, orientation, and density, as illustrated in these stylized browser tracks based on actual data. When applied to a time course of induction (e.g., up-regulated gene, A; left) or inhibition (e.g., down-regulated gene, C; right), GRO-seq can be used to determine the leading edge (left) or lagging edge (right) of the “wave” of RNA polymerase transcribing along the body of a gene. Using the edge of the RNA polymerase wave, the distance traveled over time can be measured, allowing calculation of the rate of transcription, as described [39].
Figure 7. The logic of a mitogenic transcriptional response
Estrogen signaling has immediate and lasting effects on the transcriptome in breast cancer cells, as indicated. The immediate responses prepare the cell for secondary signaling and transcriptional responses. The later responses support ribosome biogenesis, protein synthesis, and protein processing, preparing newly transcribed mRNAs for translation.
Similar articles
- Emerging Roles of Estrogen-Regulated Enhancer and Long Non-Coding RNAs.
Sedano MJ, Harrison AL, Zilaie M, Das C, Choudhari R, Ramos E, Gadad SS. Sedano MJ, et al. Int J Mol Sci. 2020 May 25;21(10):3711. doi: 10.3390/ijms21103711. Int J Mol Sci. 2020. PMID: 32466143 Free PMC article. Review. - Characterization of basal and estrogen-regulated antisense transcription in breast cancer cells: Role in regulating sense transcription.
Hou TY, Nandu T, Li R, Chae M, Murakami S, Kraus WL. Hou TY, et al. Mol Cell Endocrinol. 2020 Apr 15;506:110746. doi: 10.1016/j.mce.2020.110746. Epub 2020 Feb 5. Mol Cell Endocrinol. 2020. PMID: 32035111 Free PMC article. - Nuclear and extranuclear pathway inputs in the regulation of global gene expression by estrogen receptors.
Madak-Erdogan Z, Kieser KJ, Kim SH, Komm B, Katzenellenbogen JA, Katzenellenbogen BS. Madak-Erdogan Z, et al. Mol Endocrinol. 2008 Sep;22(9):2116-27. doi: 10.1210/me.2008-0059. Epub 2008 Jul 10. Mol Endocrinol. 2008. PMID: 18617595 Free PMC article. - Epigenomics-based identification of oestrogen-regulated long noncoding RNAs in ER+ breast cancer.
Zhang Z, Yu W, Tang D, Zhou Y, Bi M, Wang H, Zheng Y, Chen M, Li L, Xu X, Zhang W, Tao H, Jin VX, Liu Z, Chen L. Zhang Z, et al. RNA Biol. 2020 Nov;17(11):1590-1602. doi: 10.1080/15476286.2020.1777769. Epub 2020 Jun 18. RNA Biol. 2020. PMID: 32506995 Free PMC article. - Functional Relationships between Long Non-Coding RNAs and Estrogen Receptor Alpha: A New Frontier in Hormone-Responsive Breast Cancer Management.
Melone V, Salvati A, Brusco N, Alexandrova E, D'Agostino Y, Palumbo D, Palo L, Terenzi I, Nassa G, Rizzo F, Giurato G, Weisz A, Tarallo R. Melone V, et al. Int J Mol Sci. 2023 Jan 6;24(2):1145. doi: 10.3390/ijms24021145. Int J Mol Sci. 2023. PMID: 36674656 Free PMC article. Review.
Cited by
- Novel estrogen-responsive genes (ERGs) for the evaluation of estrogenic activity.
Nishi K, Fu W, Kiyama R. Nishi K, et al. PLoS One. 2022 Aug 17;17(8):e0273164. doi: 10.1371/journal.pone.0273164. eCollection 2022. PLoS One. 2022. PMID: 35976950 Free PMC article. - Deciphering direct and indirect influence of thyroid hormone with mouse genetics.
Picou F, Fauquier T, Chatonnet F, Richard S, Flamant F. Picou F, et al. Mol Endocrinol. 2014 Apr;28(4):429-41. doi: 10.1210/me.2013-1414. Epub 2014 Mar 10. Mol Endocrinol. 2014. PMID: 24617548 Free PMC article. Review. - CXXC5 is a ubiquitinated protein and is degraded by the ubiquitin-proteasome pathway.
Ayten H, Toker P, Turan Duman G, Olgun ÇE, Demiralay ÖD, Bınarcı B, Güpür G, Yaşar P, Akman HB, Haberkant P, Muyan M. Ayten H, et al. Protein Sci. 2025 Jun;34(6):e70140. doi: 10.1002/pro.70140. Protein Sci. 2025. PMID: 40371716 Free PMC article. - A PreSTIGEous use of LncRNAs to predict enhancers.
Murakami S, Gadad SS, Kraus WL. Murakami S, et al. Cell Cycle. 2015;14(11):1619-20. doi: 10.1080/15384101.2015.1032650. Cell Cycle. 2015. PMID: 25928187 Free PMC article. No abstract available. - Integrative approaches based on genomic techniques in the functional studies on enhancers.
Wang Q, Zhang J, Liu Z, Duan Y, Li C. Wang Q, et al. Brief Bioinform. 2023 Nov 22;25(1):bbad442. doi: 10.1093/bib/bbad442. Brief Bioinform. 2023. PMID: 38048082 Free PMC article. Review.
References
- Cadenas C, Bolt HM. Estrogen receptors in human disease. Arch Toxicol. 2012;86:1489–90. - PubMed
- Bjornstrom L, Sjoberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19:833–42. - PubMed
- Welboren WJ, Sweep FC, Span PN, Stunnenberg HG. Genomic actions of estrogen receptor alpha: what are the targets and how are they regulated? Endocr Relat Cancer. 2009;16:1073–89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical