Double deficiency of cathepsins B and L results in massive secretome alterations and suggests a degradative cathepsin-MMP axis - PubMed (original) (raw)
Comparative Study
Double deficiency of cathepsins B and L results in massive secretome alterations and suggests a degradative cathepsin-MMP axis
Stefan Tholen et al. Cell Mol Life Sci. 2014 Mar.
Abstract
Endolysosomal cysteine cathepsins functionally cooperate. Cathepsin B (Ctsb) and L (Ctsl) double-knockout mice die 4 weeks after birth accompanied by (autophago-) lysosomal accumulations within neurons. Such accumulations are also observed in mouse embryonic fibroblasts (MEFs) deficient for Ctsb and Ctsl. Previous studies showed a strong impact of Ctsl on the MEF secretome. Here we show that Ctsb alone has only a mild influence on extracellular proteome composition. Protease cleavage sites dependent on Ctsb were identified by terminal amine isotopic labeling of substrates (TAILS), revealing a prominent yet mostly indirect impact on the extracellular proteolytic cleavages. To investigate the cooperation of Ctsb and Ctsl, we performed a quantitative secretome comparison of wild-type MEFs and Ctsb (-/-) Ctsl (-/-) MEFs. Deletion of both cathepsins led to drastic alterations in secretome composition, highlighting cooperative functionality. While many protein levels were decreased, immunodetection corroborated increased levels of matrix metalloproteinase (MMP)-2. Re-expression of Ctsl rescues MMP-2 abundance. Ctsl and to a much lesser extent Ctsb are able to degrade MMP-2 at acidic and neutral pH. Addition of active MMP-2 to the MEF secretome degrades proteins whose levels were also decreased by Ctsb and Ctsl double deficiency. These results suggest a degradative Ctsl-MMP-2 axis, resulting in increased MMP-2 levels upon cathepsin deficiency with subsequent degradation of secreted proteins such as collagen α-1 (I).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1
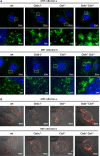
Ctsb- and Ctsl-deficient MEFs, and to a lesser extent Ctsb single and Ctsl single-deficient MEFs, show accumulations of enlarged acidic Lamp-1-positive vesicles. a Immunofluorescent staining of two wild-type MEF cell lines, two Ctsb-deficient cell lines, two Ctsl-deficient cell lines, and two Ctsb Ctsl double-deficient MEF cell lines with the lysosomal marker Lamp-1 (green). Nuclei are stained with Hoechst (blue). b Visualization of acidic vesicles with acridine orange staining
Fig. 2
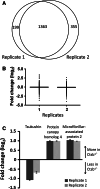
Protein identification and quantification for each biological replicate of the quantitative secretome: comparison of wild-type and Ctsb-deficient cell-conditioned media. a In both biological replicates 1,363 proteins were identified, whereas 199 and 355 proteins were found only in the first or the second replicate, respectively. b Distribution and geometric mean (horizontal bar) of fold change values (log2) of proteins from each replicate comparing the wild-type and Ctsb −_/_− MEF secretome. c Proteins with significantly altered protein abundance. These proteins are identified in both biological replicates, display an ASAPratio p value lower than 0.1 (indicated by arrow bars), show an alteration in abundance of more than 50 % (Fc < −0.58; Fc > 0.58) in both replicates (dark grey bars, replicate 1; light grey bars, replicate 2), and are annotated as secreted or extracellularly localized according to GO or Swissprot annotation
Fig. 3
N-terminal peptides identified and quantified for each biological replicate in the TAILS experiment comparing wild-type and Ctsb-deficient cell-conditioned media. a Four hundred six peptides (naturally unmodified, chemically dimethylated N-termini) were identified in both biological replicates. b Global specificity pattern upon Ctsb ablation. Graphical presentation of the secretome specificity profile of all N-termini found in the 0–20 quantile and therefore decreased upon Ctsb deletion. Only N-termini annotated as secreted or extracellularly localized according to GO or Swissprot annotation were considered. Positional occurrences are shown as enrichment over natural abundance of murine amino acid abundances as derived from the International Protein Index [74]. TAILS identifies prime-site sequences of proteolytic cleavage sites. The corresponding non-prime sequences are derived bioinformatically by database searches similar to the PICS strategy for protease specificity characterization [52, 54]
Fig. 4
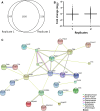
a A total of 1,826 proteins were identified in both biological replicates of the quantitative proteome comparison of wild-type and Ctsb Ctsl-deficient cell-conditioned media. b Distribution and geometric mean (horizontal bar) of fold change values (log2) of proteins from each replicate comparing the wild-type and Ctsb −_/− Ctsl −/_− MEF secretome. c Overall connectivity of all extracellular and secreted proteins altered in the quantitative proteome comparison of wild-type and Ctsb Ctsl-deficient MEFs determined by STRING (Search Tool for the Retrieval of Interacting Genes/Proteins). Different line colors represent the types of evidence for each association
Fig. 5
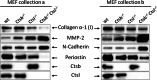
Western blot analysis of cell-conditioned media of wild-type, Ctsb-deficient, Ctsl-deficient, and Ctsb −_/− Ctsl −/_− MEFs for collagen α-1 (I), MMP-2, N-cadherin, and periostin. Absence of Ctsb and/or Ctsl was controlled by detection of the Ctsl and Ctsb proform
Fig. 6
a Western blot analysis of MMP-2 in cell-conditioned media of primary wild-type and Ctsl-deficient MEFs. b Western blot analysis of MMP-2 in cell-conditioned media of DMSO-treated wild-type MEFs and wild-type MEFs treated with 10 μM E64d for 48 h revealed an increase in MMP-2 protein levels upon cysteine cathepsin inhibition. DMSO served as solvent control. c Re-expression of Ctsl in Ctsl-deficient MEFs and overexpression in wild-type MEFs using the retroviral expression vector pMIG. Re-expression of Ctsl results in a decrease of MMP-2 protein levels. d CTSL cleaves human MMP-2 at 5.5 and 7.0. e CTSB cleaves human MMP-2 at 5.5 and 7.0, but with lower efficiency than CTSL. f Western blot analysis of cell-conditioned media of two wild-type MEF cell lines treated with activated pro-MMP-2 (125 ng/ml) compared to untreated cell-conditioned media. Pro-MMP-2 was activated by adding APMA for 3 h (details in “Materials and methods”). Collagen α-1 (I) protein levels decrease upon addition of activated pro-MMP-2 in both wild-type MEF cell lines. ON, overnight
Similar articles
- Deletion of cysteine cathepsins B or L yields differential impacts on murine skin proteome and degradome.
Tholen S, Biniossek ML, Gansz M, Gomez-Auli A, Bengsch F, Noel A, Kizhakkedathu JN, Boerries M, Busch H, Reinheckel T, Schilling O. Tholen S, et al. Mol Cell Proteomics. 2013 Mar;12(3):611-25. doi: 10.1074/mcp.M112.017962. Epub 2012 Dec 10. Mol Cell Proteomics. 2013. PMID: 23233448 Free PMC article. - Contribution of cathepsin L to secretome composition and cleavage pattern of mouse embryonic fibroblasts.
Tholen S, Biniossek ML, Gessler AL, Müller S, Weisser J, Kizhakkedathu JN, Reinheckel T, Schilling O. Tholen S, et al. Biol Chem. 2011 Nov;392(11):961-71. doi: 10.1515/BC.2011.162. Biol Chem. 2011. PMID: 21972973 - Double deficiency of cathepsin B and L in the mouse pancreas alters trypsin activity without affecting acute pancreatitis severity.
Chen W, Imasaka M, Iwama H, Nishiura H, Ohmuraya M. Chen W, et al. Pancreatology. 2022 Nov;22(7):880-886. doi: 10.1016/j.pan.2022.08.011. Epub 2022 Aug 23. Pancreatology. 2022. PMID: 36038449 - Cathepsin B Gene Knockout Improves Behavioral Deficits and Reduces Pathology in Models of Neurologic Disorders.
Hook G, Reinheckel T, Ni J, Wu Z, Kindy M, Peters C, Hook V. Hook G, et al. Pharmacol Rev. 2022 Jul;74(3):600-629. doi: 10.1124/pharmrev.121.000527. Pharmacol Rev. 2022. PMID: 35710131 Free PMC article. Review. - Recent discovery of natural substances with cathepsin L-inhibitory activity for cancer metastasis suppression.
Park JY, Park KM. Park JY, et al. Eur J Med Chem. 2024 Nov 5;277:116754. doi: 10.1016/j.ejmech.2024.116754. Epub 2024 Aug 8. Eur J Med Chem. 2024. PMID: 39128327 Review.
Cited by
- Identification of Protease Specificity by Combining Proteome-Derived Peptide Libraries and Quantitative Proteomics.
Biniossek ML, Niemer M, Maksimchuk K, Mayer B, Fuchs J, Huesgen PF, McCafferty DG, Turk B, Fritz G, Mayer J, Haecker G, Mach L, Schilling O. Biniossek ML, et al. Mol Cell Proteomics. 2016 Jul;15(7):2515-24. doi: 10.1074/mcp.O115.056671. Epub 2016 Apr 27. Mol Cell Proteomics. 2016. PMID: 27122596 Free PMC article. - Automated peptide mapping and protein-topographical annotation of proteomics data.
Videm P, Gunasekaran D, Schröder B, Mayer B, Biniossek ML, Schilling O. Videm P, et al. BMC Bioinformatics. 2014 Jun 19;15:207. doi: 10.1186/1471-2105-15-207. BMC Bioinformatics. 2014. PMID: 24946880 Free PMC article. - Cathepsin L Regulates Metabolic Networks Controlling Rapid Cell Growth and Proliferation.
Weiss-Sadan T, Itzhak G, Kaschani F, Yu Z, Mahameed M, Anaki A, Ben-Nun Y, Merquiol E, Tirosh B, Kessler B, Kaiser M, Blum G. Weiss-Sadan T, et al. Mol Cell Proteomics. 2019 Jul;18(7):1330-1344. doi: 10.1074/mcp.RA119.001392. Epub 2019 Apr 22. Mol Cell Proteomics. 2019. PMID: 31010818 Free PMC article. - Fibroblast activation protein-α, a stromal cell surface protease, shapes key features of cancer associated fibroblasts through proteome and degradome alterations.
Koczorowska MM, Tholen S, Bucher F, Lutz L, Kizhakkedathu JN, De Wever O, Wellner UF, Biniossek ML, Stahl A, Lassmann S, Schilling O. Koczorowska MM, et al. Mol Oncol. 2016 Jan;10(1):40-58. doi: 10.1016/j.molonc.2015.08.001. Epub 2015 Aug 11. Mol Oncol. 2016. PMID: 26304112 Free PMC article. - Pathomimetic cancer avatars for live-cell imaging of protease activity.
Ji K, Heyza J, Cavallo-Medved D, Sloane BF. Ji K, et al. Biochimie. 2016 Mar;122:68-76. doi: 10.1016/j.biochi.2015.09.015. Epub 2015 Sep 12. Biochimie. 2016. PMID: 26375517 Free PMC article. Review.
References
- Barbarin A, Frade R. Procathepsin L secretion, which triggers tumor progression, is regulated by Rab4A in human melanoma cells. Biochem J. 2011 - PubMed
- Joyce JA, Baruch A, Chehade K, Meyer-Morse N, Giraudo E, Tsai FY, Greenbaum DC, Hager JH, Bogyo M, Hanahan D. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5(5):443–453. doi: 10.1016/S1535-6108(04)00111-4. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous