Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells - PubMed (original) (raw)
. 2013 Aug 8;500(7461):222-6.
doi: 10.1038/nature12362. Epub 2013 Jun 30.
Kevin T Ebata, Mohammad M Karimi, Jorge A Zepeda-Martínez, Preeti Goyal, Sahasransu Mahapatra, Angela Tam, Diana J Laird, Martin Hirst, Anjana Rao, Matthew C Lorincz, Miguel Ramalho-Santos
Affiliations
- PMID: 23812591
- PMCID: PMC3893718
- DOI: 10.1038/nature12362
Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells
Kathryn Blaschke et al. Nature. 2013.
Abstract
DNA methylation is a heritable epigenetic modification involved in gene silencing, imprinting, and the suppression of retrotransposons. Global DNA demethylation occurs in the early embryo and the germ line, and may be mediated by Tet (ten eleven translocation) enzymes, which convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC). Tet enzymes have been studied extensively in mouse embryonic stem (ES) cells, which are generally cultured in the absence of vitamin C, a potential cofactor for Fe(II) 2-oxoglutarate dioxygenase enzymes such as Tet enzymes. Here we report that addition of vitamin C to mouse ES cells promotes Tet activity, leading to a rapid and global increase in 5hmC. This is followed by DNA demethylation of many gene promoters and upregulation of demethylated germline genes. Tet1 binding is enriched near the transcription start site of genes affected by vitamin C treatment. Importantly, vitamin C, but not other antioxidants, enhances the activity of recombinant Tet1 in a biochemical assay, and the vitamin-C-induced changes in 5hmC and 5mC are entirely suppressed in Tet1 and Tet2 double knockout ES cells. Vitamin C has a stronger effect on regions that gain methylation in cultured ES cells compared to blastocysts, and in vivo are methylated only after implantation. In contrast, imprinted regions and intracisternal A particle retroelements, which are resistant to demethylation in the early embryo, are resistant to vitamin-C-induced DNA demethylation. Collectively, the results of this study establish vitamin C as a direct regulator of Tet activity and DNA methylation fidelity in ES cells.
Figures
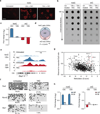
Figure 1. Vitamin C induces loss of mC at gene promoters via a transient increase in hmC
a, Immunofluorescence for hmC. Scale bar is 200 µm. b, Global hmC and mC levels assayed by dot blot. c, Graph shows fold change in DIP-seq reads (RPKM) at methylated promoters (n = 1045) for VitC-treated cells relative to untreated cells. Values are mean ± s.e.m. d, Overlap of methylated promoters that gain hmC and those that lose mC. P value calculated using Fisher’s exact test. e, Genome browser view of Dazl.f, Bisulfite sequencing of promoters. Open circles = unmethylated, closed circles = methylated. g, Scatter plot of methylated promoters comparing change in methylation (Z score) versus CpG content. Red circles = imprinted genes. h, Graphs show fold change in RPKM at retrotransposons for VitC-treated cells relative to untreated cells. Values are mean ± s.e.m.
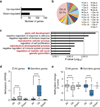
Figure 2. Vitamin C-induced DNA demethylation leads to expression of germline genes
a, Number of genes differentially expressed following VitC treatment (2-fold and P < 0.05 by t-test). b, Chromosomal distribution of up-regulated genes. c, Gene ontology analysis of up-regulated genes. d, Box plot showing basal promoter methylation levels (RPKM) in untreated ESCs for all genes on the microarray (n = 18,023), up-regulated genes (n = 102), down-regulated genes (n = 48), all germline genes (n = 865), up-regulated germline genes (n = 8), and down-regulated germline genes (n = 3). e, Box plot showing the extent of VitC-induced demethylation (Z score) at gene promoters categorized as in panel d. The box plots have Tukey whiskers, a line for the median, and edges for the 25th/75th percentile. ****P < 0.0001 by ANOVA throughout the figure.
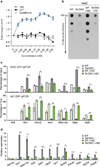
Figure 3. The effects of Vitamin C are Tet-dependent
a, Dose-dependent effect of VitC on in vitro Tet activity. n = 3 technical replicates, values are mean ± s.d. b, Dot blot for hmC following a 12 hr VitC treatment. c, hmC (top) and mC (bottom) DIP-qPCR following a 12 or 72 hr VitC treatment, respectively. d, Gene expression at 72 hrs of VitC treatment. qRT-PCR data expressed relative to untreated wild-type cells. For c, d, n = 2 biological replicates, values are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001 by t-test throughout the figure.
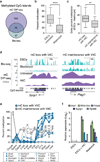
Figure 4. Vitamin C reduces DNA methylation in ESCs that is normally gained post-implantation
a, Overlap of methylated CGIs in ESCs from this study (mC DIP-seq, RPKM >0.5) and ref. (whole genome bisulfite (bis)-seq, >25% methylated CpGs). Only CGIs found to be methylated in both data sets were used for subsequent analysis in b–d. b, The box plot shows CGI methylation levels in ESCs and blastocysts (values from ref. , ****P < 0.0001 by t-test). **c**, CGIs were first categorized as having mC loss with VitC (>75% loss of mC at 72 hrs, n = 23) or mC maintenance with VitC (<25% loss of mC at 72 hrs, n = 14) in ESCs, then plotted for difference in methylation between untreated ESCs and blastocysts. CGIs demethylated upon VitC treatment show significantly greater ESC hypermethylation compared to CGIs resistant to VitC (**P < 0.01 by t-test). d, Genome browser views of a gene from each category described in panel c. e, Methylation levels during development of all genes categorized in panel c for which data exist (ref. 2). f, Gene expression in ESCs cultured with or without VitC compared to E3.5 blastocysts. Data expressed relative to untreated ESCs. n = 2 biological replicates, values are mean ± s.e.m.
Similar articles
- PRDM14 promotes active DNA demethylation through the ten-eleven translocation (TET)-mediated base excision repair pathway in embryonic stem cells.
Okashita N, Kumaki Y, Ebi K, Nishi M, Okamoto Y, Nakayama M, Hashimoto S, Nakamura T, Sugasawa K, Kojima N, Takada T, Okano M, Seki Y. Okashita N, et al. Development. 2014 Jan;141(2):269-80. doi: 10.1242/dev.099622. Epub 2013 Dec 11. Development. 2014. PMID: 24335252 - Ascorbate-induced generation of 5-hydroxymethylcytosine is unaffected by varying levels of iron and 2-oxoglutarate.
Dickson KM, Gustafson CB, Young JI, Züchner S, Wang G. Dickson KM, et al. Biochem Biophys Res Commun. 2013 Oct 4;439(4):522-7. doi: 10.1016/j.bbrc.2013.09.010. Epub 2013 Sep 8. Biochem Biophys Res Commun. 2013. PMID: 24021282 Free PMC article. - Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification.
Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Ito S, et al. Nature. 2010 Aug 26;466(7310):1129-33. doi: 10.1038/nature09303. Nature. 2010. PMID: 20639862 Free PMC article. - Tet family of 5-methylcytosine dioxygenases in mammalian development.
Zhao H, Chen T. Zhao H, et al. J Hum Genet. 2013 Jul;58(7):421-7. doi: 10.1038/jhg.2013.63. Epub 2013 May 30. J Hum Genet. 2013. PMID: 23719188 Free PMC article. Review. - DNA methylation: TET proteins-guardians of CpG islands?
Williams K, Christensen J, Helin K. Williams K, et al. EMBO Rep. 2011 Dec 23;13(1):28-35. doi: 10.1038/embor.2011.233. EMBO Rep. 2011. PMID: 22157888 Free PMC article. Review.
Cited by
- A distal Foxp3 enhancer enables interleukin-2 dependent thymic Treg cell lineage commitment for robust immune tolerance.
Dikiy S, Li J, Bai L, Jiang M, Janke L, Zong X, Hao X, Hoyos B, Wang ZM, Xu B, Fan Y, Rudensky AY, Feng Y. Dikiy S, et al. Immunity. 2021 May 11;54(5):931-946.e11. doi: 10.1016/j.immuni.2021.03.020. Epub 2021 Apr 9. Immunity. 2021. PMID: 33838102 Free PMC article. - High-Dose Vitamin C in Advanced-Stage Cancer Patients.
Zasowska-Nowak A, Nowak PJ, Ciałkowska-Rysz A. Zasowska-Nowak A, et al. Nutrients. 2021 Feb 26;13(3):735. doi: 10.3390/nu13030735. Nutrients. 2021. PMID: 33652579 Free PMC article. Review. - Ascorbate as a co-factor for fe- and 2-oxoglutarate dependent dioxygenases: physiological activity in tumor growth and progression.
Kuiper C, Vissers MC. Kuiper C, et al. Front Oncol. 2014 Dec 10;4:359. doi: 10.3389/fonc.2014.00359. eCollection 2014. Front Oncol. 2014. PMID: 25540771 Free PMC article. Review. - Ascorbate content of clinical glioma tissues is related to tumour grade and to global levels of 5-hydroxymethyl cytosine.
Crake RLI, Burgess ER, Wiggins GAR, Magon NJ, Das AB, Vissers MCM, Morrin HR, Royds JA, Slatter TL, Robinson BA, Phillips E, Dachs GU. Crake RLI, et al. Sci Rep. 2022 Sep 1;12(1):14845. doi: 10.1038/s41598-022-19032-8. Sci Rep. 2022. PMID: 36050369 Free PMC article. - High-Dose Vitamin C: Preclinical Evidence for Tailoring Treatment in Cancer Patients.
Giansanti M, Karimi T, Faraoni I, Graziani G. Giansanti M, et al. Cancers (Basel). 2021 Mar 20;13(6):1428. doi: 10.3390/cancers13061428. Cancers (Basel). 2021. PMID: 33804775 Free PMC article. Review.
References
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Development. 2002;16:6–21. - PubMed
- Gu T-P, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 92093/CAPMC/ CIHR/Canada
- HD065812/HD/NICHD NIH HHS/United States
- R01 AI044432/AI/NIAID NIH HHS/United States
- R01 HD065812/HD/NICHD NIH HHS/United States
- R01 CA151535/CA/NCI NIH HHS/United States
- CA151535/CA/NCI NIH HHS/United States
- DP2OD004698/OD/NIH HHS/United States
- R01 OD012204/OD/NIH HHS/United States
- DP2 OD004698/OD/NIH HHS/United States
- P30 DK063720/DK/NIDDK NIH HHS/United States
- DP2 OD007420/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases