Tracking a refined eIF4E-binding motif reveals Angel1 as a new partner of eIF4E - PubMed (original) (raw)
Tracking a refined eIF4E-binding motif reveals Angel1 as a new partner of eIF4E
Pauline Gosselin et al. Nucleic Acids Res. 2013 Sep.
Abstract
The initiation factor 4E (eIF4E) is implicated in most of the crucial steps of the mRNA life cycle and is recognized as a pivotal protein in gene regulation. Many of these roles are mediated by its interaction with specific proteins generally known as eIF4E-interacting partners (4E-IPs), such as eIF4G and 4E-BP. To screen for new 4E-IPs, we developed a novel approach based on structural, in silico and biochemical analyses. We identified the protein Angel1, a member of the CCR4 deadenylase family. Immunoprecipitation experiments provided evidence that Angel1 is able to interact in vitro and in vivo with eIF4E. Point mutation variants of Angel1 demonstrated that the interaction of Angel1 with eIF4E is mediated through a consensus eIF4E-binding motif. Immunofluorescence and cell fractionation experiments showed that Angel1 is confined to the endoplasmic reticulum and Golgi apparatus, where it partially co-localizes with eIF4E and eIF4G, but not with 4E-BP. Furthermore, manipulating Angel1 levels in living cells had no effect on global translation rates, suggesting that the protein has a more specific function. Taken together, our results illustrate that we developed a powerful method for identifying new eIF4E partners and open new perspectives for understanding eIF4E-specific regulation.
Figures
Figure 1.
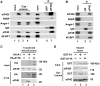
New screening reveals a novel 4E-IP, Angel1, which acquired an eIF4E-binding motif in vertebrates. (A) Combination of structural, in silico and m7GTP chromatography approaches reveal that Angel1 is a novel eIF4E-interacting protein. See text for details. (B) The putative eIF4E-binding motifs of Angel1 mouse (m), human (h), chicken (Ga) and Xenopus (Xe) were aligned over several eIF4E-binding proteins. Residues that were identical or conserved in >75% of the sequences are shaded in black and gray, respectively. The seven last amino acids correspond to the consensus eIF4E-binding motif YxxxxLΦ. (C) Unrooted phylogenetic tree of Angel-related sequences in 9 species. The presented tree was constructed using the Maximum Likelihood method (see SI). The brace indicates the sequences that contain the consensus motif (YxxxxLΦ).
Figure 2.
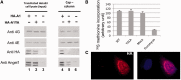
Angel1 interacts with eIF4E through its eIF4E-binding motif. (A, B) One HeLa S3 cell extract supplemented with RNase A was incubated with m7GTP beads or used to perform immunoprecipitation with indicated antibodies. Bound proteins were analyzed by western blotting. (A) Sepharose beads or m7GTP beads (cap-column assay) were incubated directly with Laemmli buffer (lanes 1 and 2). m7GTP beads were incubated with 200 µM free m7GTP; eluates (lane 4) and residual proteins attached to m7GTP beads (lane 3) were resolved on SDS-PAGE. Immunoprecipitations with anti-eIF4GI (lane 7) or isotype control antibody (anti-GFP, lane 6) contain immunoglobulin (IgG). Total lysate is presented in lane 5. (B) Immunoprecipitation using anti-eIF4E (lane 3) or isotype control antibody (anti-HA, lane 2) were performed with the same total lysate (lane 1) as in A. (C) Angel1 interacts with eIF4E through the conserved eIF4E-binding sequence. HeLa S3 cells were mock-transfected, or transfected with HA-A1 or HA-A1YA expressing vectors. Cell lysates were subjected to HA-immunoprecipitation. Whole lysates (input) and immnoprecipitates (IP HA) were analyzed by immunoblotting. (D) Angel1 competes with eIF4GI for binding to eIF4E in vitro. HeLa S3 cell lysates were supplemented with wild-type (lane 2) or mutant Angel1 (lane 3) GST-fusion protein (1 µg each) and incubated with m7GTP beads. 1/50 of total extracts (Input) and proteins bound to m7GTP beads (cap-column) were analyzed by western blot. The ability of recombinant wild-type or mutant Angel1 to bind endogenous eIF4E and displace eIF4GI was monitored using an anti-GST antibody.
Figure 3.
Overexpressed Angel1 neither competes with eIF4G nor affects general translation activity. HeLa S3 cells were mock-transfected, or transfected with HA-A1 or HA-A1YA expressing vectors. (A) Transfected cells were lysed and used for m7GTP chromatography and analyzed by immunoblotting. The membrane incubated with the anti-HA tag was then reprobed with the anti-Angel1 antibody. (B) In parallel, 24 h after transfection, cells were incubated with 35S-methionine and treated as described in ref. using TCA to precipitate labeled proteins. 35S-methionine incorporation into proteins is expressed as a percentage of the mock-transfected control (n = 5). (C) Localization of HA-Angel1 was determined by indirect immunofluorescence with anti-HA and anti-Rat IgG-TRITC antibodies. Nuclei were stained with 1 µg/ml of Hoescht.
Figure 4.
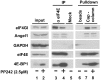
Angel1–eIF4E interaction is not sensitive to mTOR inhibition. HeLa S3 cells were treated with or without 2.5 µM PP242 (mTOR inhibitor) for 1 h. Cell extracts were incubated with m7GTP beads (cap-column, lanes 7 and 8), α-eIF4E-sepharose beads (lanes 3 and 4) or sepharose beads alone (lanes 5 and 6), as described in Figure 2A and B. Whole cell lysates (lanes 1 and 2) and bound proteins were analyzed by immunoblotting.
Figure 5.
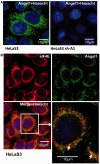
Angel1 co-localizes specifically with eIF4E in small perinuclear granules. (A) Immunofluorescence staining was performed with the anti-Angel1 antibody (Sigma) and an Alexa 488-conjugated anti-rabbit secondary antibody (green) on HeLa S3 cells or sh-Angel1 expressing cell lines. Nuclei were stained with 1 µg/ml of Hoescht. The subcellular localization of Angel1 was visualized using confocal microscopy. (B) Immunofluorescence staining was performed on HeLa S3 cells with the anti-Angel1 and Alexa 488-conjugated anti-rabbit (green) and an anti-eIF4E-specific polyclonal Alexa 555-conjugated antibody (red). Co-localization of Angel1 and eIF4E appears in yellow and is indicated by white arrows.
Figure 6.
Angel1 is co-distributed with the ER and the Golgi apparatus. Co-localization of Angel1 and Calnexin (A), or GM130 (B) was determined by indirect immunofluorescence with respectively anti-Angel1 and Alexa 488-conjugated anti-rabbit (green), anti-Calnexin and Cy3-conjugated anti-mouse (red) or anti-GM130 and Cy3-conjugated anti-mouse antibodies (red). Subcellular localization of Angel1, Calnexin and GM130 were visualized using confocal microscopy. Co-localization between Angel1 and Calnexin (A), and Angel1 and GM130 (B) appears in yellow on the merged images (right panels). (C) Angel1 co-fractionated with Golgi and ER elements. HeLa S3 cells underwent subcellular fractionation. Nuclear (N), microsomal (M), perinuclear (P) and cytosolic fractions (C) were analyzed by western blotting with the indicated antibodies.
Similar articles
- Mitosis-related phosphorylation of the eukaryotic translation suppressor 4E-BP1 and its interaction with eukaryotic translation initiation factor 4E (eIF4E).
Sun R, Cheng E, Velásquez C, Chang Y, Moore PS. Sun R, et al. J Biol Chem. 2019 Aug 2;294(31):11840-11852. doi: 10.1074/jbc.RA119.008512. Epub 2019 Jun 14. J Biol Chem. 2019. PMID: 31201269 Free PMC article. - The 4E-BP Caf20p Mediates Both eIF4E-Dependent and Independent Repression of Translation.
Castelli LM, Talavera D, Kershaw CJ, Mohammad-Qureshi SS, Costello JL, Rowe W, Sims PF, Grant CM, Hubbard SJ, Ashe MP, Pavitt GD. Castelli LM, et al. PLoS Genet. 2015 May 14;11(5):e1005233. doi: 10.1371/journal.pgen.1005233. eCollection 2015 May. PLoS Genet. 2015. PMID: 25973932 Free PMC article. - The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation.
Grüner S, Peter D, Weber R, Wohlbold L, Chung MY, Weichenrieder O, Valkov E, Igreja C, Izaurralde E. Grüner S, et al. Mol Cell. 2016 Nov 3;64(3):467-479. doi: 10.1016/j.molcel.2016.09.020. Epub 2016 Oct 20. Mol Cell. 2016. PMID: 27773676 - Control of the eIF4E activity: structural insights and pharmacological implications.
Romagnoli A, D'Agostino M, Ardiccioni C, Maracci C, Motta S, La Teana A, Di Marino D. Romagnoli A, et al. Cell Mol Life Sci. 2021 Nov;78(21-22):6869-6885. doi: 10.1007/s00018-021-03938-z. Epub 2021 Sep 19. Cell Mol Life Sci. 2021. PMID: 34541613 Free PMC article. Review. - Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation?
Scheper GC, Proud CG. Scheper GC, et al. Eur J Biochem. 2002 Nov;269(22):5350-9. doi: 10.1046/j.1432-1033.2002.03291.x. Eur J Biochem. 2002. PMID: 12423333 Free PMC article. Review.
Cited by
- BTG4 is a meiotic cell cycle-coupled maternal-zygotic-transition licensing factor in oocytes.
Yu C, Ji SY, Sha QQ, Dang Y, Zhou JJ, Zhang YL, Liu Y, Wang ZW, Hu B, Sun QY, Sun SC, Tang F, Fan HY. Yu C, et al. Nat Struct Mol Biol. 2016 May;23(5):387-94. doi: 10.1038/nsmb.3204. Epub 2016 Apr 11. Nat Struct Mol Biol. 2016. PMID: 27065194 - eIF4E3 forms an active eIF4F complex during stresses (eIF4FS) targeting mTOR and re-programs the translatome.
Weiss B, Allen GE, Kloehn J, Abid K, Jaquier-Gubler P, Curran JA. Weiss B, et al. Nucleic Acids Res. 2021 May 21;49(9):5159-5176. doi: 10.1093/nar/gkab267. Nucleic Acids Res. 2021. PMID: 33893802 Free PMC article. - Activity-dependent neuroprotective protein (ADNP) exhibits striking sexual dichotomy impacting on autistic and Alzheimer's pathologies.
Malishkevich A, Amram N, Hacohen-Kleiman G, Magen I, Giladi E, Gozes I. Malishkevich A, et al. Transl Psychiatry. 2015 Feb 3;5(2):e501. doi: 10.1038/tp.2014.138. Transl Psychiatry. 2015. PMID: 25646590 Free PMC article. - ANGEL2 phosphatase activity is required for non-canonical mitochondrial RNA processing.
Clemente P, Calvo-Garrido J, Pearce SF, Schober FA, Shigematsu M, Siira SJ, Laine I, Spåhr H, Steinmetzger C, Petzold K, Kirino Y, Wibom R, Rackham O, Filipovska A, Rorbach J, Freyer C, Wredenberg A. Clemente P, et al. Nat Commun. 2022 Sep 30;13(1):5750. doi: 10.1038/s41467-022-33368-9. Nat Commun. 2022. PMID: 36180430 Free PMC article. - hu.MAP3.0: Atlas of human protein complexes by integration of > 25,000 proteomic experiments.
Fischer SN, Claussen ER, Kourtis S, Sdelci S, Orchard S, Hermjakob H, Kustatscher G, Drew K. Fischer SN, et al. bioRxiv [Preprint]. 2024 Oct 15:2024.10.11.617930. doi: 10.1101/2024.10.11.617930. bioRxiv. 2024. PMID: 39464102 Free PMC article. Preprint.
References
- Strudwick S, Borden KL. The emerging roles of translation factor eIF4E in the nucleus. Differentiation. 2002;70:10–22. - PubMed
- von der Haar T, Gross JD, Wagner G, McCarthy JE. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 2004;11:503–511. - PubMed
- Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. - PubMed
- Clemens MJ. Targets and mechanisms for the regulation of translation in malignant transformation. Oncogene. 2004;23:3180–3188. - PubMed
- Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–750. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous