Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA - PubMed (original) (raw)
Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA
Lakshminarayan M Iyer et al. Nucleic Acids Res. 2013 Sep.
Abstract
Discovery of the TET/JBP family of dioxygenases that modify bases in DNA has sparked considerable interest in novel DNA base modifications and their biological roles. Using sensitive sequence and structure analyses combined with contextual information from comparative genomics, we computationally characterize over 12 novel biochemical systems for DNA modifications. We predict previously unidentified enzymes, such as the kinetoplastid J-base generating glycosyltransferase (and its homolog GREB1), the catalytic specificity of bacteriophage TET/JBP proteins and their role in complex DNA base modifications. We also predict the enzymes involved in synthesis of hypermodified bases such as alpha-glutamylthymine and alpha-putrescinylthymine that have remained enigmatic for several decades. Moreover, the current analysis suggests that bacteriophages and certain nucleo-cytoplasmic large DNA viruses contain an unexpectedly diverse range of DNA modification systems, in addition to those using previously characterized enzymes such as Dam, Dcm, TET/JBP, pyrimidine hydroxymethylases, Mom and glycosyltransferases. These include enzymes generating modified bases such as deazaguanines related to queuine and archaeosine, pyrimidines comparable with lysidine, those derived using modified S-adenosyl methionine derivatives and those using TET/JBP-generated hydroxymethyl pyrimidines as biosynthetic starting points. We present evidence that some of these modification systems are also widely dispersed across prokaryotes and certain eukaryotes such as basidiomycetes, chlorophyte and stramenopile alga, where they could serve as novel epigenetic marks for regulation or discrimination of self from non-self DNA. Our study extends the role of the PUA-like fold domains in recognition of modified nucleic acids and predicts versions of the ASCH and EVE domains to be novel 'readers' of modified bases in DNA. These results open opportunities for the investigation of the biology of these systems and their use in biotechnology.
Figures
Figure 1.
Pathways are numbered in the order of discussion in the ‘Results and Discussion’ section. Names of enzymes are given above reaction arrows. Location of the reaction in DNA or free bases is indicated in parentheses below reaction arrows. Chemical groups added during a step are highlighted in red in the ensuing product. Green labels indicate entry points for pathway intermediates and products into non-base modification pathways. The orange dot indicates a compound and the purple dot a DNA-modification enzyme predicted here for the first time.
Figure 2.
Previously described and well-supported eukaryotic clades of TET/JBP proteins are collapsed. Nodes supported by bootstrap >75% are shown. Relevant domain architectures and operons of TET/JBP and TAGT families are shown between or below the trees. Proteins are either denoted by a species abbreviation (eukaryotes and bacteria) or by the complete name (phages/viruses) followed by GenBank GIs. Branch coloring: Phage sequences- blue, bacteria- pink, kinetoplastid- red and _C. subellipsoidea_- green. See
Supplementary Data
for species abbreviations.
Figure 3.
Multiple sequence alignment of the DNA base glycosyltransferases (A) and the aG/P-T-pyrophosphorylases (B). Protein sequences are labeled by gene names followed by species abbreviation and Genbank GIs. Phage protein names are colored blue, bacterial ones in pink, archaea in orange and eukaryotes in black. Predicted catalytic residues for both TAGT and aG/PT-PPlase are indicated by asterisks, with secondary structure assignments shown above the alignment. Alignment columns are colored based on the 80% conservation consensus. Topology of the glycosyltransferase domain is adjacent to the alignment. See
Supplementary Data
for species abbreviations.
Figure 4.
Gene neighborhoods and contextual information network of domains involved in DNA modification. Genes are shown as arrows pointing from the 5′ to the 3′ end. Representative gene-neighborhoods are labeled with taxonomic lineage and species name of origin and an anchor GI number of the gene marked with an asterisk. Operons are grouped by the type of predicted DNA modification they catalyze and also by Roman numerals to match corresponding nodes in the network. Domain architectures are shown as insets. In the network, gray edges connect neighboring genes (5′–>3′), and magenta-colored edges connect neighboring domains (N–>C terminus). Nodes are clustered either by their common function or by the predicted DNA modifications they catalyze. Thickness of edges reflects the frequency of associations between two nodes. The graph was calculated from 1751 gene neighborhoods, including 101 nodes, 377 pairwise connections and 5922 genes.
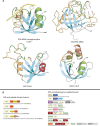
Figure 5.
Structures, domain architectures and operons of representative members of the PUA-like fold. (A) Structures are centered on the conserved binding cleft with regions contributing to it colored in green. Family-specific inserts are colored in orange. The EVE structure was crystallized with a 3[N-morpholino]propane sulfonic acid molecule in the predicted binding site. (B). Genes in operons are represented and labeled as described in Figure 4. Fusions of the EVE-like domain are shown to the left.
Similar articles
- Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids.
Iyer LM, Tahiliani M, Rao A, Aravind L. Iyer LM, et al. Cell Cycle. 2009 Jun 1;8(11):1698-710. doi: 10.4161/cc.8.11.8580. Epub 2009 Jun 27. Cell Cycle. 2009. PMID: 19411852 Free PMC article. - Lineage-specific expansions of TET/JBP genes and a new class of DNA transposons shape fungal genomic and epigenetic landscapes.
Iyer LM, Zhang D, de Souza RF, Pukkila PJ, Rao A, Aravind L. Iyer LM, et al. Proc Natl Acad Sci U S A. 2014 Feb 4;111(5):1676-83. doi: 10.1073/pnas.1321818111. Epub 2014 Jan 7. Proc Natl Acad Sci U S A. 2014. PMID: 24398522 Free PMC article. - Phage-encoded ten-eleven translocation dioxygenase (TET) is active in C5-cytosine hypermodification in DNA.
Burke EJ, Rodda SS, Lund SR, Sun Z, Zeroka MR, O'Toole KH, Parker MJ, Doshi DS, Guan C, Lee YJ, Dai N, Hough DM, Shnider DA, Corrêa IR Jr, Weigele PR, Saleh L. Burke EJ, et al. Proc Natl Acad Sci U S A. 2021 Jun 29;118(26):e2026742118. doi: 10.1073/pnas.2026742118. Proc Natl Acad Sci U S A. 2021. PMID: 34155108 Free PMC article. - Adenine methylation in eukaryotes: Apprehending the complex evolutionary history and functional potential of an epigenetic modification.
Iyer LM, Zhang D, Aravind L. Iyer LM, et al. Bioessays. 2016 Jan;38(1):27-40. doi: 10.1002/bies.201500104. Epub 2015 Dec 12. Bioessays. 2016. PMID: 26660621 Free PMC article. Review. - Introduction: Metals in Biology: α-Ketoglutarate/Iron-Dependent Dioxygenases.
Guengerich FP. Guengerich FP. J Biol Chem. 2015 Aug 21;290(34):20700-20701. doi: 10.1074/jbc.R115.675652. Epub 2015 Jul 7. J Biol Chem. 2015. PMID: 26152720 Free PMC article. Review.
Cited by
- CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems.
Makarova KS, Anantharaman V, Grishin NV, Koonin EV, Aravind L. Makarova KS, et al. Front Genet. 2014 Apr 30;5:102. doi: 10.3389/fgene.2014.00102. eCollection 2014. Front Genet. 2014. PMID: 24817877 Free PMC article. - Comprehensive classification of the PIN domain-like superfamily.
Matelska D, Steczkiewicz K, Ginalski K. Matelska D, et al. Nucleic Acids Res. 2017 Jul 7;45(12):6995-7020. doi: 10.1093/nar/gkx494. Nucleic Acids Res. 2017. PMID: 28575517 Free PMC article. - Switching transcription with bacterial RNA polymerase through photocaging, photorelease and phosphorylation reactions in the major groove of DNA.
Vaníková Z, Janoušková M, Kambová M, Krásný L, Hocek M. Vaníková Z, et al. Chem Sci. 2019 Mar 4;10(14):3937-3942. doi: 10.1039/c9sc00205g. eCollection 2019 Apr 14. Chem Sci. 2019. PMID: 31015933 Free PMC article. - Dissecting the HGT network of carbon metabolic genes in soil-borne microbiota.
Li L, Liu Y, Xiao Q, Xiao Z, Meng D, Yang Z, Deng W, Yin H, Liu Z. Li L, et al. Front Microbiol. 2023 Jul 7;14:1173748. doi: 10.3389/fmicb.2023.1173748. eCollection 2023. Front Microbiol. 2023. PMID: 37485539 Free PMC article. - Identification of the glucosyltransferase that converts hydroxymethyluracil to base J in the trypanosomatid genome.
Bullard W, Lopes da Rosa-Spiegler J, Liu S, Wang Y, Sabatini R. Bullard W, et al. J Biol Chem. 2014 Jul 18;289(29):20273-82. doi: 10.1074/jbc.M114.579821. Epub 2014 Jun 2. J Biol Chem. 2014. PMID: 24891501 Free PMC article.
References
- Bloomfield VA, Crothers DM, Tinomo IJ. Nucleic Acids: Structures, Properties and Functions. Sausalito, CA: University Science Books; 2000.
- Gommers-Ampt JH, Borst P. Hypermodified bases in DNA. FASEB J. 1995;9:1034–1042. - PubMed
- Iyer LM, Abhiman S, Aravind L. Natural history of eukaryotic DNA methylation systems. Prog. Mol. Biol. Transl. Sci. 2011;101:25–104. - PubMed
- Warren RA. Modified bases in bacteriophage DNAs. Annu. Rev. Microbiol. 1980;34:137–158. - PubMed
- Greenberg GR, He P, Hilfinger J, Tseng MJ. In: Molecular Biology of Bacteriophage T4. Karam JD, editor. Washington, DC: American Society of Microbiology; 1994. pp. 14–27.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases