From human genome to cancer genome: the first decade - PubMed (original) (raw)
Review
From human genome to cancer genome: the first decade
David A Wheeler et al. Genome Res. 2013 Jul.
Abstract
The realization that cancer progression required the participation of cellular genes provided one of several key rationales, in 1986, for embarking on the human genome project. Only with a reference genome sequence could the full spectrum of somatic changes leading to cancer be understood. Since its completion in 2003, the human reference genome sequence has fulfilled its promise as a foundational tool to illuminate the pathogenesis of cancer. Herein, we review the key historical milestones in cancer genomics since the completion of the genome, and some of the novel discoveries that are shaping our current understanding of cancer.
Figures
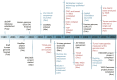
Figure 1.
Major events in a decade of cancer genomics. (Dark blue) Major advances in massively parallel sequencing platforms and targeted enrichment technologies; (black) major large-scale projects designed to catalog genomic variations of normal human individuals; (red) cancer genomics. (dbSNP) Database of single nucleotide polymorphism; (HapMap) haplotype map of the human genome; (ENCODE) Encyclopedia of DNA Elements; (COSMIC) Catalog of Somatic Mutations in Cancer; (TCGA) The Cancer Genome Atlas; (GA) genome analyzer; (CRC) colorectal carcinoma; (WES) whole-exome sequencing; (ICGC) International Cancer Genome Consortium; (TSP) tumor sequencing project; (AML) acute myeloid leukemia; (WGS) whole-genome sequencing; (OSCC) ovarian small cell carcinoma.
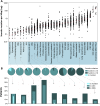
Figure 2.
Frequencies of somatic mutations in cancer patients. All data represents primary tumors. Only nonsilent mutations (missense, nonsense, frameshift, and splice site) were counted. (A) Overall frequencies of somatic mutations. Each black dot represents a tumor. The light blue shaded group indicates pediatric tumors, and the deeper blue shaded group indicates adult tumors. Red horizontal lines within each cluster of points indicate median value of the mutation frequency of each tumor type. (ALL) Acute lymphoblastic leukemia; (AML) acute myeloid leukemia; (C) carcinoma; (GCT) germ cell tumor; (CRC) colorectal carcinoma; (MSI) microsatellite instability; (MSS) microsatellite stable; (POLE) patients with somatic mutation in the nuclease (proofreading) domain of the POLE gene. The outlier in the low-grade glioma patient with >100 mutations per Mb is also _POLE_-mutated. (B) Frequency classification of tumors. The pie charts divide the patients into three groups based on frequency of nonsilent mutation: 0 detectable somatic mutations, less than 30, and greater than or equal to 30 for selected representative tumor types (30 mutations represent a frequency of 1 per Mbp in A). The nested histograms below the pie charts show the percentage of patients with no significantly mutated genes (SMG, calculated by MutSig, q ≤ 0.1), no cancer census genes (CGC), or no mutations at all. The sequencing data for all the pediatric tumors, CRC, and hepatocellular carcinoma were generated at the Human Genome Sequencing Center at Baylor College of Medicine. The sequencing data for all other adult tumors were from the TCGA Genome Data Analysis Center (
https://confluence.broadinstitute.org/display/GDAC/Home
). Pediatric AML, ALL, and Wilm's Tumor data were obtained from the TARGET project (
).
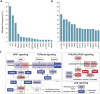
Figure 3.
Significantly mutated genes and principal cancer pathways deregulated by somatic mutations in human colorectal carcinoma. Patients are divided into two groups based on mutation rate. All genes shown are significantly mutated with a false discovery rate of less than 0.1. (A) Profile determined from 193 patients with chromosome instable, low mutation rate, disease (see Fig. 2A, CRC MSS). (B) Profile determined from 29 microsatellite instable CRC plus 7 _POLE_-mutated patients (see Fig. 2A, CRC MSI and CRC MSS POLE). (C) Principal cancer pathways deregulated by somatic mutation in CRC. Alterations are defined by somatic mutations, homozygous deletions, high-level focal amplifications, and, in some cases, by significant up- or down-regulation of gene expression (black up-triangle). All genes from Figure 3 except MLK4, GPC6, and EDNRB can be placed in one of the four pathways shown here. WNT signaling is disrupted by one or more mutations in 93% of patients; TGFbeta signaling is disrupted in 26% of all patients with a low mutation rate and in 94% of patients, and RTK/RAS/PI3K signaling is disrupted in over 80% of patients. (Red) Activated genes; (blue) inactivated genes. Deep red or blue are genes on the significantly mutated list from panels A and B. Lighter shaded genes are not mutated significantly in this cohort but contribute to pathway disruption in some patients. Panels A and B adapted from Figure 1, and panel C from Figure 4, of The Cancer Genome Atlas Research Network (2012a).
Similar articles
- Big science: The cancer genome challenge.
Ledford H. Ledford H. Nature. 2010 Apr 15;464(7291):972-4. doi: 10.1038/464972a. Nature. 2010. PMID: 20393534 No abstract available. - Cancer genome landscapes.
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Vogelstein B, et al. Science. 2013 Mar 29;339(6127):1546-58. doi: 10.1126/science.1235122. Science. 2013. PMID: 23539594 Free PMC article. Review. - Our changing view of the genomic landscape of cancer.
Bell DW. Bell DW. J Pathol. 2010 Jan;220(2):231-43. doi: 10.1002/path.2645. J Pathol. 2010. PMID: 19918804 Free PMC article. Review. - Genetic and epigenetic heterogeneity in cancer: a genome-centric perspective.
Heng HH, Bremer SW, Stevens JB, Ye KJ, Liu G, Ye CJ. Heng HH, et al. J Cell Physiol. 2009 Sep;220(3):538-47. doi: 10.1002/jcp.21799. J Cell Physiol. 2009. PMID: 19441078 Review. - Decade in review--genomics: a decade of discovery in cancer genomics.
Offit K. Offit K. Nat Rev Clin Oncol. 2014 Nov;11(11):632-4. doi: 10.1038/nrclinonc.2014.170. Epub 2014 Oct 7. Nat Rev Clin Oncol. 2014. PMID: 25286975 No abstract available.
Cited by
- Differences in genome-wide repeat sequence instability conferred by proofreading and mismatch repair defects.
Lujan SA, Clark AB, Kunkel TA. Lujan SA, et al. Nucleic Acids Res. 2015 Apr 30;43(8):4067-74. doi: 10.1093/nar/gkv271. Epub 2015 Mar 30. Nucleic Acids Res. 2015. PMID: 25824945 Free PMC article. - Ploidy-Seq: inferring mutational chronology by sequencing polyploid tumor subpopulations.
Malhotra A, Wang Y, Waters J, Chen K, Meric-Bernstam F, Hall IM, Navin NE. Malhotra A, et al. Genome Med. 2015 Jan 28;7(1):6. doi: 10.1186/s13073-015-0127-5. eCollection 2015. Genome Med. 2015. PMID: 25729435 Free PMC article. - Tailored Antibacterials and Innovative Laboratories for Phage (Φ) Research: Personalized Infectious Disease Medicine for the Most Vulnerable At-Risk Patients.
Terwilliger AL, Gu Liu C, Green SI, Clark JR, Salazar KC, Hernandez Santos H, Heckmann ER, Trautner BW, Ramig RF, Maresso AW. Terwilliger AL, et al. Phage (New Rochelle). 2020 Jun 1;1(2):66-74. doi: 10.1089/phage.2020.0007. Epub 2020 Jun 16. Phage (New Rochelle). 2020. PMID: 32626851 Free PMC article. - Multi-Omics Approaches in Colorectal Cancer Screening and Diagnosis, Recent Updates and Future Perspectives.
Ullah I, Yang L, Yin FT, Sun Y, Li XH, Li J, Wang XJ. Ullah I, et al. Cancers (Basel). 2022 Nov 11;14(22):5545. doi: 10.3390/cancers14225545. Cancers (Basel). 2022. PMID: 36428637 Free PMC article. Review. - Genome-wide analysis of gynecologic cancer: The Cancer Genome Atlas in ovarian and endometrial cancer.
Iijima M, Banno K, Okawa R, Yanokura M, Iida M, Takeda T, Kunitomi-Irie H, Adachi M, Nakamura K, Umene K, Nogami Y, Masuda K, Tominaga E, Aoki D. Iijima M, et al. Oncol Lett. 2017 Mar;13(3):1063-1070. doi: 10.3892/ol.2017.5582. Epub 2017 Jan 10. Oncol Lett. 2017. PMID: 28454214 Free PMC article.
References
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. 2007. Direct selection of human genomic loci by microarray hybridization. Nat Methods 4: 903–905 - PubMed
- Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, Kempski H, Moorman AV, Titley I, Swansbury J, et al. 2011. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469: 356–361 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources