Principles and concepts of DNA replication in bacteria, archaea, and eukarya - PubMed (original) (raw)
Review
Principles and concepts of DNA replication in bacteria, archaea, and eukarya
Michael O'Donnell et al. Cold Spring Harb Perspect Biol. 2013.
Abstract
The accurate copying of genetic information in the double helix of DNA is essential for inheritance of traits that define the phenotype of cells and the organism. The core machineries that copy DNA are conserved in all three domains of life: bacteria, archaea, and eukaryotes. This article outlines the general nature of the DNA replication machinery, but also points out important and key differences. The most complex organisms, eukaryotes, have to coordinate the initiation of DNA replication from many origins in each genome and impose regulation that maintains genomic integrity, not only for the sake of each cell, but for the organism as a whole. In addition, DNA replication in eukaryotes needs to be coordinated with inheritance of chromatin, developmental patterning of tissues, and cell division to ensure that the genome replicates once per cell division cycle.
Figures
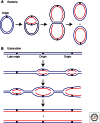
Figure 1.
Replication initiation in bacteria and eukaryotes. (A) Most bacteria have a circular chromosome with one origin, although there are exceptions to this. Illustrated here is the E. coli chromosome that has one origin from which two replication forks proceed in opposite directions. (B) Eukaryotes have long linear chromosomes. Bidirectional replication is initiated at multiple origins along each chromosome.
Figure 2.
Origin activation and replisome assembly in bacteria and eukaryotes. (A) Origin activation in eukaryotes is regulated by DDK (Dbf4-Cdc7 kinase) and CDK (cyclin-dependent protein kinase) kinases that have low activity in G1 phase, and high activity in S phase. (B) Steps in origin activation and replisome assembly in bacteria and eukaryotes. The relatively more complex process of DNA replication in eukaryotes is reflected in the larger number of proteins required to initiate and elongate DNA synthesis from each origin. See text for details.
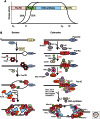
Figure 3.
Organization of bacterial and eukaryotic replisome machines. (A) Replisome architecture in E. coli. The helicase (DnaB) encircles the lagging strand. Three molecules of Pol III are attached to one clamp loader. The clamp loader binds the helicase and repeatedly assembles β clamps onto primed sites as they are formed by primase. (B) Proposed architecture of a eukaryotic replisome. The MCM2-7 helicase encircles the leading strand; unwinding is aided by association of GINS and Cdc45 with MCM2-7 to form the CMG complex. The RFC clamp loader repeatedly loads PCNA (proliferating cell nuclear antigen) clamps onto lagging-strand primers formed by Pol α-primase. Unlike E. coli, the clamp loader may not form stabile attachments to the replisome. The leading-strand polymerase (Pol ε) is stabilized on DNA by Mrc1. Pol δ replicates the lagging strand. Contacts between Pol δ and other components of the replisome are not yet defined. Mcm10 and Ctf4 contact Pol α-primase.
Similar articles
- Why are there so many diverse replication machineries?
Forterre P. Forterre P. J Mol Biol. 2013 Nov 29;425(23):4714-26. doi: 10.1016/j.jmb.2013.09.032. Epub 2013 Sep 26. J Mol Biol. 2013. PMID: 24075868 Review. - Common domains in the initiators of DNA replication in Bacteria, Archaea and Eukarya: combined structural, functional and phylogenetic perspectives.
Giraldo R. Giraldo R. FEMS Microbiol Rev. 2003 Jan;26(5):533-54. doi: 10.1111/j.1574-6976.2003.tb00629.x. FEMS Microbiol Rev. 2003. PMID: 12586394 Review. - Evolution and origin of sliding clamp in bacteria, archaea and eukarya.
Acharya S, Dahal A, Bhattarai HK. Acharya S, et al. PLoS One. 2021 Aug 11;16(8):e0241093. doi: 10.1371/journal.pone.0241093. eCollection 2021. PLoS One. 2021. PMID: 34379636 Free PMC article. - Origins of DNA replication in the three domains of life.
Robinson NP, Bell SD. Robinson NP, et al. FEBS J. 2005 Aug;272(15):3757-66. doi: 10.1111/j.1742-4658.2005.04768.x. FEBS J. 2005. PMID: 16045748 Review. - The replication machinery of LUCA: common origin of DNA replication and transcription.
Koonin EV, Krupovic M, Ishino S, Ishino Y. Koonin EV, et al. BMC Biol. 2020 Jun 9;18(1):61. doi: 10.1186/s12915-020-00800-9. BMC Biol. 2020. PMID: 32517760 Free PMC article.
Cited by
- Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation.
Georgescu RE, Schauer GD, Yao NY, Langston LD, Yurieva O, Zhang D, Finkelstein J, O'Donnell ME. Georgescu RE, et al. Elife. 2015 Apr 14;4:e04988. doi: 10.7554/eLife.04988. Elife. 2015. PMID: 25871847 Free PMC article. - Orc1 Binding to Mitotic Chromosomes Precedes Spatial Patterning during G1 Phase and Assembly of the Origin Recognition Complex in Human Cells.
Kara N, Hossain M, Prasanth SG, Stillman B. Kara N, et al. J Biol Chem. 2015 May 8;290(19):12355-69. doi: 10.1074/jbc.M114.625012. Epub 2015 Mar 17. J Biol Chem. 2015. PMID: 25784553 Free PMC article. - High-density lipoprotein regulates angiogenesis by long non-coding RNA HDRACA.
Mo ZW, Peng YM, Zhang YX, Li Y, Kang BA, Chen YT, Li L, Sorci-Thomas MG, Lin YJ, Cao Y, Chen S, Liu ZL, Gao JJ, Huang ZP, Zhou JG, Wang M, Chang GQ, Deng MJ, Liu YJ, Ma ZS, Hu ZJ, Dong YG, Ou ZJ, Ou JS. Mo ZW, et al. Signal Transduct Target Ther. 2023 Aug 14;8(1):299. doi: 10.1038/s41392-023-01558-6. Signal Transduct Target Ther. 2023. PMID: 37574469 Free PMC article. - Identification of PCNA-interacting protein motifs in human DNA polymerase δ.
Khandagale P, Thakur S, Acharya N. Khandagale P, et al. Biosci Rep. 2020 Apr 30;40(4):BSR20200602. doi: 10.1042/BSR20200602. Biosci Rep. 2020. PMID: 32314787 Free PMC article. - The interaction networks of the budding yeast and human DNA replication-initiation proteins.
Wu R, Amin A, Wang Z, Huang Y, Man-Hei Cheung M, Yu Z, Yang W, Liang C. Wu R, et al. Cell Cycle. 2019 Mar-Apr;18(6-7):723-741. doi: 10.1080/15384101.2019.1586509. Epub 2019 Mar 19. Cell Cycle. 2019. PMID: 30890025 Free PMC article.
References
- Araki H 2010. Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr Opin Cell Biol 22: 766–771 - PubMed
Publication types
MeSH terms
Grants and funding
- R37 GM038839/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P30 CA045508/CA/NCI NIH HHS/United States
- P01 CA013106/CA/NCI NIH HHS/United States
- R01 GM038839/GM/NIGMS NIH HHS/United States
- CA13016/CA/NCI NIH HHS/United States
- GM38839/GM/NIGMS NIH HHS/United States
- GM45436/GM/NIGMS NIH HHS/United States
- R01 GM045436/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources