TET1 plays an essential oncogenic role in MLL-rearranged leukemia - PubMed (original) (raw)
. 2013 Jul 16;110(29):11994-9.
doi: 10.1073/pnas.1310656110. Epub 2013 Jul 1.
Xi Jiang, Zejuan Li, Yuanyuan Li, Chun-Xiao Song, Chunjiang He, Miao Sun, Ping Chen, Sandeep Gurbuxani, Jiapeng Wang, Gia-Ming Hong, Abdel G Elkahloun, Stephen Arnovitz, Jinhua Wang, Keith Szulwach, Li Lin, Craig Street, Mark Wunderlich, Meelad Dawlaty, Mary Beth Neilly, Rudolf Jaenisch, Feng-Chun Yang, James C Mulloy, Peng Jin, Paul P Liu, Janet D Rowley, Mingjiang Xu, Chuan He, Jianjun Chen
Affiliations
- PMID: 23818607
- PMCID: PMC3718141
- DOI: 10.1073/pnas.1310656110
TET1 plays an essential oncogenic role in MLL-rearranged leukemia
Hao Huang et al. Proc Natl Acad Sci U S A. 2013.
Abstract
The ten-eleven translocation 1 (TET1) gene is the founding member of the TET family of enzymes (TET1/2/3) that convert 5-methylcytosine to 5-hydroxymethylcytosine. Although TET1 was first identified as a fusion partner of the mixed lineage leukemia (MLL) gene in acute myeloid leukemia carrying t(10,11), its definitive role in leukemia is unclear. In contrast to the frequent down-regulation (or loss-of-function mutations) and critical tumor-suppressor roles of the three TET genes observed in various types of cancers, here we show that TET1 is a direct target of MLL-fusion proteins and is significantly up-regulated in MLL-rearranged leukemia, leading to a global increase of 5-hydroxymethylcytosine level. Furthermore, our both in vitro and in vivo functional studies demonstrate that Tet1 plays an indispensable oncogenic role in the development of MLL-rearranged leukemia, through coordination with MLL-fusion proteins in regulating their critical cotargets, including homeobox A9 (Hoxa9)/myeloid ecotropic viral integration 1 (Meis1)/pre-B-cell leukemia homeobox 3 (Pbx3) genes. Collectively, our data delineate an MLL-fusion/Tet1/Hoxa9/Meis1/Pbx3 signaling axis in MLL-rearranged leukemia and highlight TET1 as a potential therapeutic target in treating this presently therapy-resistant disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
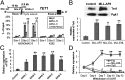
Fig. 1.
TET1 is aberrantly overexpressed in _MLL_-rearranged AML and is likely a direct target of MLL and particularly MLL-fusions. (A_–_C) Relative expression levels of TET1 (A), TET2 (B), and TET3 (C) in 12 _MLL_-rearranged AML, 88 non–_MLL_-rearranged AML, and nine normal control samples as detected by microarrays. The average expression level of each TET gene in normal controls was set as 1. Note that the gene expression values used in our analyses were log-transformed, not the original absolute values (
SI Materials and Methods
). (D) Relative expression levels of TET1 as detected by qPCR. The average level of TET1 expression in CD33+ cell samples was set as 1. MLL, _MLL_-rearranged; NC, normal control; MNC, mononuclear cells.
Fig. 2.
TET1 is a direct target of MLL and particularly MLL-fusions. (A) ChIP-qPCR assays of the enrichment of MLL-N (i.e., MLL N-terminal, representing both wild-type MLL and MLL-fusion proteins), MLL-C (i.e., MLL C-terminal, representing wild-type MLL only), and H3K79me2 at the promoter region of TET1 (sites 2 and 3) and a distal upstream region (site 1) in MONOMAC-6 and K562 cells. IgG was used as a negative control. (B) qPCR (Lower) and Western blotting (Upper) analyses of Tet1 expression in mouse normal BM progenitor (Lin−) cells that were transduced with MSCV-MLL-AF9, -MLL-ELL, -MLL-ENL, or empty vector (Control). Transduced cells were cultured in methylcellulose for 7 d. (C) qPCR analysis of TET1 expression in human cord blood CD34+ cells that were transduced with MSCV-MLL-AF9 or empty vector and cultured for a period to select transduction-positive cells (MA9-1 to -5, represent different lines of immortalized cells) (35). (D) qPCR analysis of MLL-ENL, Tet1, or Fas expression in mouse MLL-ENL-ERtm cells after withdrawal of 4-OHT. MA9, MLL-AF9. *P < 0.05; **P < 0.01, two-tailed t test. Pgk1, phosphoglycerate kinase 1.
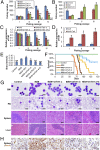
Fig. 3.
Effects of Tet1 in MLL-AF9 (MA9)-mediated cell transformation and leukemogenesis. (A and B) Effects of depletion (A) or forced expression (B) of Tet1 on _MA9_-mediated cell transformation. (C and D) qPCR analysis of Tet1 expression in different passages of colony cells shown in A and B. (E) Dot blot assay of 5hmC level in genomic DNA of colony cells (passage II) shown in A and B. A similar pattern was observed in liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) assays. (F) Kaplan–Meier survival analysis of the BM transplantation recipient mice. The median survival of MA9, MA9+shTet1-a, MA9+shTet1-b, and MA9+shTet1-a+b mice (n = 10 for each group) is 70, 85, 108, and over 150 d, respectively; MA9+shTet1-a vs. MA9, P = 0.003; MA9+shTet1-b vs. MA9, P = 0.0003; MA9+shTet1-a+b vs. MA9, P < 0.00001; log-rank test. (G) Wright–Giemsa-stained BM cell cytospin and peripheral blood (PB) smear, and hematoxylin/eosin (H&E)-stained spleen and liver paraffin sections of transplantation recipient mice are shown. (Scale bars: 10 μm in BM/ PB and 100 μm in spleen/liver.) (H) 5hmC antibody-stained spleen. (Scale bars: 20 μm.) *P < 0.05; **P < 0.01, two-tailed t test; in A_–_E, MA9 samples were used as controls for the statistical comparisons. Note: “MA9” represents “MSCVneo-MA9+pGFP-V-RS-scrambled shRNA” in all of the plots except for B and D where it represents “MSCVneo-MA9+MSCVpuro”; “Control” represents “MSCVneo+MSCVpuro” or “MSCVneo+pGFP-V-RS” (see
SI Materials and Methods
for more details).
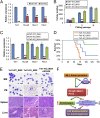
Fig. 4.
HOXA9, MEIS1, and PBX3 are important targets of TET1. (A_–_C) ChIP-qPCR assay of the binding of TET1, as well as MLL-fusion proteins, to the loci of HOXA9 (A), MEIS1 (B), and PBX3 (C). Green bars represent CpG islands. Brown-purple bars represent exons of target genes. (D and E) The effects of knockdown of TET1 by siRNA oligos (D, Left, qPCR; Right, Western blot**)** and of ectopic expression of mouse Tet1 (E) on expression of TET1, HOXA9, MEIS1, and PBX3 in human MONOMAC-6 leukemic cells. (F and G) Effects of knockdown of Tet1 by different shRNAs on expression of the four genes in colony cells (F) or in BM cells of transplanted mice (G, Left, qPCR; Right, Western blot**)**; see Fig. 3 for more details about the samples. (H and I) Effects of knockdown TET1 by siRNAs with or without cotransfection of HOXA9, MEIS1, or PBX3 on apoptosis (H, Left), cell viability (H, Right), and cell growth/proliferation (I) of _MLL_-rearranged leukemic cells. siNC, scrambled siRNA oligos (as negative control of siTet1); +H (+M, +P, or +T) or +HOXA9 (+MEIS1, +PBX3, or +Tet1), cotransfected with MSCVpuro-HOXA9 (-MEIS1, -PBX3, or -Tet1). PGK1/Pgk1 was used as endogenous control for qPCR. *P < 0.05; **P < 0.01, t test; in A_–_C, K562 was used as a control for MONOMAC-6 for statistics analysis of each item. In G, MA9 group was used as the control.
Fig. 5.
Effects of _Tet1_-knockout and the signaling-pathway model. (A) Expression changes of three target genes in Tet1 knockout BM progenitor (i.e., Lin−) cells relative to wild-type controls. (B and C) Effect of Tet1 knockout on MA9-mediated cell transformation and the corresponding expression changes of the targets. (D) Kaplan–Meier survival analysis of the BM transplantation recipient mice. The median survival of Tet1-WT_MA9 (MSCVneo-MA9+MSCVpuro cotransduced into wild-type mouse BM progenitor cells; n = 8), Tet1-KO_MA9 (MSCVneo-MA9+MSCVpuro cotransduced into Tet1−/− mouse BM progenitor cells; n = 9), and Tet1-KO_MA9+HOXA9 (MSCVneo-MA9+MSCVpuro-HOXA9 cotransduced into Tet1−/− mouse BM progenitor cells; n = 5) is 66, >150, and 85 d, respectively; Tet1-WT_MA9 vs. Tet1-KO_MA9, P < 0.00001; Tet1-WT_MA9 vs. Tet1-KO_MA9+HOXA9, P = 0.018; Tet1-KO_MA9 vs. Tet1-KO_MA9+HOXA9, P = 0.019; log-rank test. (E) Wright–Giemsa-stained BM cell cytospin and PB smear, and hematoxylin/eosin (H&E)-stained spleen and liver paraffin sections of transplantation recipient mice are shown. (Scales bars: 10 μm in BM/PB and 100 μm in spleen/liver.) (F) The model of the MLL-fusion/Tet1/Hoxa9/Meis1/Pbx3 signaling axis in _MLL_-rearranged leukemia. *P < 0.05; **P < 0.01, t test.
Similar articles
- Identification of MLL-fusion/MYC⊣miR-26⊣TET1 signaling circuit in MLL-rearranged leukemia.
Huang H, Jiang X, Wang J, Li Y, Song CX, Chen P, Li S, Gurbuxani S, Arnovitz S, Wang Y, Weng H, Neilly MB, He C, Li Z, Chen J. Huang H, et al. Cancer Lett. 2016 Mar 28;372(2):157-65. doi: 10.1016/j.canlet.2015.12.032. Epub 2016 Jan 11. Cancer Lett. 2016. PMID: 26791235 Free PMC article. - PBX3 and MEIS1 Cooperate in Hematopoietic Cells to Drive Acute Myeloid Leukemias Characterized by a Core Transcriptome of the MLL-Rearranged Disease.
Li Z, Chen P, Su R, Hu C, Li Y, Elkahloun AG, Zuo Z, Gurbuxani S, Arnovitz S, Weng H, Wang Y, Li S, Huang H, Neilly MB, Wang GG, Jiang X, Liu PP, Jin J, Chen J. Li Z, et al. Cancer Res. 2016 Feb 1;76(3):619-29. doi: 10.1158/0008-5472.CAN-15-1566. Epub 2016 Jan 8. Cancer Res. 2016. PMID: 26747896 Free PMC article. - Initiation of MLL-rearranged AML is dependent on C/EBPα.
Ohlsson E, Hasemann MS, Willer A, Lauridsen FK, Rapin N, Jendholm J, Porse BT. Ohlsson E, et al. J Exp Med. 2014 Jan 13;211(1):5-13. doi: 10.1084/jem.20130932. Epub 2013 Dec 23. J Exp Med. 2014. PMID: 24367003 Free PMC article. - Deregulation of the HOXA9/MEIS1 axis in acute leukemia.
Collins CT, Hess JL. Collins CT, et al. Curr Opin Hematol. 2016 Jul;23(4):354-61. doi: 10.1097/MOH.0000000000000245. Curr Opin Hematol. 2016. PMID: 27258906 Free PMC article. Review. - Loss of 5-hydroxymethylcytosine in cancer: cause or consequence?
Ficz G, Gribben JG. Ficz G, et al. Genomics. 2014 Nov;104(5):352-7. doi: 10.1016/j.ygeno.2014.08.017. Epub 2014 Aug 30. Genomics. 2014. PMID: 25179374 Free PMC article. Review.
Cited by
- A pan-cancer analysis of MYC-PVT1 reveals CNV-unmediated deregulation and poor prognosis in renal carcinoma.
Posa I, Carvalho S, Tavares J, Grosso AR. Posa I, et al. Oncotarget. 2016 Jul 26;7(30):47033-47041. doi: 10.18632/oncotarget.9487. Oncotarget. 2016. PMID: 27366943 Free PMC article. - Prolyl hydroxylase domain enzymes: important regulators of cancer metabolism.
Yang M, Su H, Soga T, Kranc KR, Pollard PJ. Yang M, et al. Hypoxia (Auckl). 2014 Aug 30;2:127-142. doi: 10.2147/HP.S47968. eCollection 2014. Hypoxia (Auckl). 2014. PMID: 27774472 Free PMC article. Review. - Making it or breaking it: DNA methylation and genome integrity.
Sriraman A, Debnath TK, Xhemalce B, Miller KM. Sriraman A, et al. Essays Biochem. 2020 Oct 26;64(5):687-703. doi: 10.1042/EBC20200009. Essays Biochem. 2020. PMID: 32808652 Free PMC article. Review. - Epigenetic control of adult stem cell function.
Avgustinova A, Benitah SA. Avgustinova A, et al. Nat Rev Mol Cell Biol. 2016 Oct;17(10):643-58. doi: 10.1038/nrm.2016.76. Epub 2016 Jul 13. Nat Rev Mol Cell Biol. 2016. PMID: 27405257 Review. - Hydroxymethylation as a Novel Environmental Biosensor.
Dao T, Cheng RY, Revelo MP, Mitzner W, Tang W. Dao T, et al. Curr Environ Health Rep. 2014 Mar 1;1(1):1-10. doi: 10.1007/s40572-013-0005-5. Curr Environ Health Rep. 2014. PMID: 24860723 Free PMC article.
References
- Gu TP, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477(7366):606–610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA118319/CA/NCI NIH HHS/United States
- CA127277/CA/NCI NIH HHS/United States
- R37 HD045022/HD/NICHD NIH HHS/United States
- HG006827/HG/NHGRI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- HL112294/HL/NHLBI NIH HHS/United States
- R01 HG006827/HG/NHGRI NIH HHS/United States
- R01 NS079625/NS/NINDS NIH HHS/United States
- HD073162/HD/NICHD NIH HHS/United States
- R21 HD073162/HD/NICHD NIH HHS/United States
- R01 CA178454/CA/NCI NIH HHS/United States
- R01 CA127277/CA/NCI NIH HHS/United States
- NS079625/NS/NINDS NIH HHS/United States
- R01 HL112294/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials