Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system - PubMed (original) (raw)
Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system
Andrew R Bassett et al. Cell Rep. 2013.
Erratum in
- Highly Efficient Targeted Mutagenesis of Drosophila with the CRISPR/Cas9 System.
Bassett AR, Tibbit C, Ponting CP, Liu JL. Bassett AR, et al. Cell Rep. 2014 Mar 27;6(6):1178-1179. doi: 10.1016/j.celrep.2014.03.017. Epub 2014 Mar 27. Cell Rep. 2014. PMID: 28898684 Free PMC article. No abstract available.
Abstract
Here, we present a simple and highly efficient method for generating and detecting mutations of any gene in Drosophila melanogaster through the use of the CRISPR/Cas9 system (clustered regularly interspaced palindromic repeats/CRISPR-associated). We show that injection of RNA into the Drosophila embryo can induce highly efficient mutagenesis of desired target genes in up to 88% of injected flies. These mutations can be transmitted through the germline to make stable lines. Our system provides at least a 10-fold improvement in efficiency over previously published reports, enabling wider application of this technique. We also describe a simple and highly sensitive method of detecting mutations in the target gene by high-resolution melt analysis and discuss how the new technology enables the study of gene function.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Graphical abstract
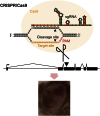
Figure 1
CRISPR/Cas9 System for Targeting Double-Strand Breaks (A) The two-component system for inducing double-strand breaks. The synthetic guide RNA (sgRNA) contains a region of complementarity to the target site on the DNA, as well as stem loops from the tracrRNA to mediate binding to the Cas9 protein. Cas9 protein is indicated by a yellow circle, cleavage sites by arrowheads, and the protospacer adjacent motif (PAM, NGG) required for cleavage in red. (B) The PCR-based system for generation of sgRNAs. Two oligonucleotides are used to generate the sgRNA template for in vitro transcription (black lines). The first includes a T7 promoter (highlighted in blue) and upstream sequence for efficient in vitro transcription, followed by a GGN18 target-site sequence and a portion of the sgRNA stem loops. The second includes the entire sgRNA sequence after the target site. PCR is performed with the two primers but without any other DNA template. See also Figure S1 and Table S1.
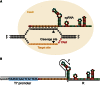
Figure 2
CRISPR/Cas9-Induced Deletions at the yellow and white Loci (A) A schematic of the yellow and white genes showing the sgRNA target sites. Exons are shown as black boxes, transcriptional start sites as arrows, and sgRNA target sites as black triangles. (B) Sequence of sgRNA targets. The 20 nt target sequence corresponding to each target site is indicated in orange, along with the neighboring NGG protospacer adjacent motif (PAM) in red. (C) Mosaic yellow expression in the injected G0 flies. Female flies showing mosaic yellow expression upon injection with Cas9 mRNA and y1 sgRNA are shown. Patches of yellow mutant tissue can be observed (arrowheads) and are outlined by dotted lines. (D) Mosaic white expression in the injected G0 flies. Female and male flies showing mosaic white expression upon injection with Cas9 mRNA and w2 sgRNA are shown. Patches of white mutant tissue can be observed (arrowheads). (E) Sequencing of induced mutations in the yellow gene. PCR products spanning the sgRNA target sites were analyzed for indels. The first line in each alignment represents wild-type sequence, and subsequent lines show individual mutant clones. Target sites are indicated in orange, PAM in red, and cleavage sites by a black triangle. Deleted bases are marked with dashes, and inserted or substituted bases are indicated in lower case. (F) Sequencing of induced mutations in the white gene, as in (E).
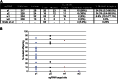
Figure 3
Germline Transmission of Mutations (A) Efficiency of germline transmission of mutations induced at each target site. Number of embryos injected, hatched L1 larvae, and total and visibly mosaic G0 adults produced are noted. Each G0 adult was crossed to flies with yellow and white gene mutations (_y_1_w_1), and the number of flies producing offspring was noted (fertile), along with the number of crosses that produced at least one mutant offspring (germline mutants). This is also expressed as a percentage of the total G0 adults (brackets). The percentage of mutant progeny shows the total number of mutant offspring compared to the total number of offspring from all crosses. The range shown in brackets shows the percentage of mutant flies produced from each individual positive cross. The bottom row of the table shows the equivalent numbers from Gratz et al. (2013) for comparison. (B) Percentage of mutant offspring from each injected G0 fly. Offspring of each injected G0 fly were backcrossed to flies with mutations in yellow and white genes (_y_1_w_1) and analyzed for the percentage of yellow or white mutant female offspring. Each individual cross is represented by a single point. Distributions for each target site are shown, and they ranged from 0% to 88.5%.
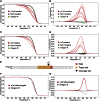
Figure 4
High-Resolution Melt Analysis (A) High resolution melt analysis (HRMA) of G0 mosaic yellow mutant flies. Mosaic mutant flies (red) can be easily differentiated from wild-type Oregon R (green) and nonmosaic (gray) flies by a change in the shape of the melt curve due to heteroduplex formation. (B) Change in fluorescence relative to control Oregon R and nonmosaic flies highlight the changes in curve shape shown in (A). (C) HRMA performed as in (A) but for white mosaic flies. (D) Change in fluorescence as in (B) but for white mosaic flies. (E) Sequencing of the y1a mutant shows a single point deletion compared to wild-type Oregon R flies at the y1 cleavage site. Target sites are indicated in orange, the protospacer adjacent motif (PAM) in red, and the cleavage site by a black triangle. (F) HRMA shows a reproducible shift in the melting curve in a heterozygous point deletion in the yellow gene (y1a, red) in comparison to wild-type Oregon R flies (gray). (G) Change in fluorescence highlights the difference in curve shape shown in (F).
Figure 5
Analysis of Off-Target Effects by High-Resolution Melt Analysis (A) High-resolution melt analysis (HRMA) of the putative off-target site in the CG14073 gene in y1 sgRNA-injected mosaic flies. The upper panel shows the alignment of the off-target site with the y1 sgRNA sequence, indicating the target site in orange, PAM sequence in red, and “seed” sequence previously shown to be required for target recognition (Cong et al., 2013) with a black box. The lower panel shows HRMA of the putative off-target site, showing no difference between wild-type Oregon R flies (gray) and mosaic yellow mutant flies (colored). (B) HRMA performed as in (A) but for y2 sgRNA off-target sites in CG43672 and H4r. (C) HRMA performed as in (A) but for w1 sgRNA off-target sites in CG13397 and Myo10A. (D) HRMA performed as in (A) but for w2 sgRNA off-target sites in CG34422 and Fim.
Figure S1
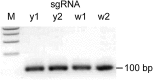
sgRNA Template PCR, Related to Figure 1 Representative gel image of sgRNA template PCR. 5% of the reaction from each sgRNA template PCR was analyzed on a 1.5% agarose gel. A single band of the expected size is observed. M – 1 kb ladder (New England Biolabs).
Similar articles
- Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease.
Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O'Connor-Giles KM. Gratz SJ, et al. Genetics. 2013 Aug;194(4):1029-35. doi: 10.1534/genetics.113.152710. Epub 2013 May 24. Genetics. 2013. PMID: 23709638 Free PMC article. - CRISPR/Cas9 mediated genome engineering in Drosophila.
Bassett A, Liu JL. Bassett A, et al. Methods. 2014 Sep;69(2):128-36. doi: 10.1016/j.ymeth.2014.02.019. Epub 2014 Feb 24. Methods. 2014. PMID: 24576617 - CRISPR-Directed Gene Editing Catalyzes Precise Gene Segment Replacement In Vitro Enabling a Novel Method for Multiplex Site-Directed Mutagenesis.
Sansbury BM, Wagner AM, Tarcic G, Barth S, Nitzan E, Goldfus R, Vidne M, Kmiec EB. Sansbury BM, et al. CRISPR J. 2019 Apr;2:121-132. doi: 10.1089/crispr.2018.0054. CRISPR J. 2019. PMID: 30998096 - CRISPR/Cas9 and genome editing in Drosophila.
Bassett AR, Liu JL. Bassett AR, et al. J Genet Genomics. 2014 Jan 20;41(1):7-19. doi: 10.1016/j.jgg.2013.12.004. Epub 2013 Dec 18. J Genet Genomics. 2014. PMID: 24480743 Review. - A Toolkit of CRISPR-Based Genome Editing Systems in Drosophila.
Xu J, Ren X, Sun J, Wang X, Qiao HH, Xu BW, Liu LP, Ni JQ. Xu J, et al. J Genet Genomics. 2015 Apr 20;42(4):141-9. doi: 10.1016/j.jgg.2015.02.007. Epub 2015 Mar 12. J Genet Genomics. 2015. PMID: 25953352 Review.
Cited by
- Evaluation of CRISPR/Cas9 site-specific function and validation of sgRNA sequence by a Cas9/sgRNA-assisted reverse PCR technique.
Zhang B, Zhou J, Li M, Wei Y, Wang J, Wang Y, Shi P, Li X, Huang Z, Tang H, Song Z. Zhang B, et al. Anal Bioanal Chem. 2021 Apr;413(9):2447-2456. doi: 10.1007/s00216-021-03173-2. Epub 2021 Mar 4. Anal Bioanal Chem. 2021. PMID: 33661348 Free PMC article. - An enhanced gene targeting toolkit for Drosophila: Golic+.
Chen HM, Huang Y, Pfeiffer BD, Yao X, Lee T. Chen HM, et al. Genetics. 2015 Mar;199(3):683-94. doi: 10.1534/genetics.114.173716. Epub 2015 Jan 2. Genetics. 2015. PMID: 25555988 Free PMC article. - Advances and perspectives in the application of CRISPR/Cas9 in insects.
Chen L, Wang G, Zhu YN, Xiang H, Wang W. Chen L, et al. Dongwuxue Yanjiu. 2016 Jul 18;37(4):220-8. doi: 10.13918/j.issn.2095-8137.2016.4.220. Dongwuxue Yanjiu. 2016. PMID: 27469253 Free PMC article. Review. - Expanding CRISPR/Cas9 Genome Editing Capacity in Zebrafish Using SaCas9.
Feng Y, Chen C, Han Y, Chen Z, Lu X, Liang F, Li S, Qin W, Lin S. Feng Y, et al. G3 (Bethesda). 2016 Aug 9;6(8):2517-21. doi: 10.1534/g3.116.031914. G3 (Bethesda). 2016. PMID: 27317783 Free PMC article. - A Novel Neuroprotective Role of Phosphatase of Regenerating Liver-1 against CO2 Stimulation in Drosophila.
Guo P, Xu X, Wang F, Yuan X, Tu Y, Zhang B, Zheng H, Yu D, Ge W, Gong Z, Yang X, Xi Y. Guo P, et al. iScience. 2019 Sep 27;19:291-302. doi: 10.1016/j.isci.2019.07.026. Epub 2019 Jul 22. iScience. 2019. PMID: 31404830 Free PMC article.
References
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_U137788471/MRC_/Medical Research Council/United Kingdom
- MC_UU_12021/1/MRC_/Medical Research Council/United Kingdom
- MC_U137761446/MRC_/Medical Research Council/United Kingdom
- MC_UU_12021/3/MRC_/Medical Research Council/United Kingdom
- 249869/ERC_/European Research Council/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials