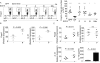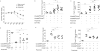The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis - PubMed (original) (raw)
The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis
Patrick M Smith et al. Science. 2013.
Abstract
Regulatory T cells (Tregs) that express the transcription factor Foxp3 are critical for regulating intestinal inflammation. Candidate microbe approaches have identified bacterial species and strain-specific molecules that can affect intestinal immune responses, including species that modulate Treg responses. Because neither all humans nor mice harbor the same bacterial strains, we posited that more prevalent factors exist that regulate the number and function of colonic Tregs. We determined that short-chain fatty acids, gut microbiota-derived bacterial fermentation products, regulate the size and function of the colonic Treg pool and protect against colitis in a Ffar2-dependent manner in mice. Our study reveals that a class of abundant microbial metabolites underlies adaptive immune microbiota coadaptation and promotes colonic homeostasis and health.
Figures
Fig. 1. SCFA restore colonic Treg populations and function in germ-free mice
(A) Colonic lamina propria (LP) lymphocytes were isolated and stained for CD4 and Foxp3. Left panel: Representative dot plots and percentage of CD4+Foxp3+ within the CD45+CD3+ population from SPF, GF, and GF mice treated with propionate (P), acetate (A), butyrate (B), or the SCFA mix in the drinking water. Right panel: Numbers of Foxp3+ Tregs for the left panel. (B) cTregs were isolated from in vivo propionate-treated GF mice, sorted by CD4, CD127, and CD25, and examined ex vivo for expression of Foxp3 and IL-10 by RTqPCR. (C) cTregs were isolated from GF mice and purified as in 1B, cultured for 24 hrs in the presence or absence of 0.1 mM propionate and examined for expression of Foxp3, TGFβ, and IL-10 by RTqPCR and IL-10 protein production by ELISA. For panel A, symbols represent data from individual mice. Horizontal lines show the mean and error bars the SD. For panels B and C, each symbol or bar represents pooled cTregs from 3-5 mice. All data shown are representative of at least 3 independent experiments. A Kruskal-Wallis test with a Dunn's post hoc test was performed in panel A, *** P <0.001 and * P <0.05. A Mann-Whitney U test was performed in panels B and C.
Fig. 2. SCFA augment colonic Treg population size and function in SPF mice
(A) SPF Foxp3YFP-Cre mice were treated with water alone (-) or P, A, B, or the mix. Colonic LP lymphocytes were isolated and stained for CD4 and IL-10. Upper panel: Representative dot plots and percentage of CD4+ Foxp3-YFP+ within the CD45+CD3+ population. Lower panel: Representative dot plots and percentage of the CD4+Foxp3+IL-10+population. (B) Cell numbers for the data in (A) upper panel. (C) Cell numbers for the data in (A) lower panel. Symbols in B and C represent data from individual mice and represent 4 independent experiments. (D) cTregs were co-cultured with splenic effector T cells (Teff) and P, A, B, or media (sodium and pH matched) for 96 hrs. Percent suppression (y-axis), Treg:Teff ratios (x-axis). Symbols represent mean values and error bars SD for four independent experiments. * P < 0.01. (E) cTregs were isolated from the LP of in vivo propionate treated SPF Foxp3YFP-Cre mice, sorted for CD4 and YFP, and examined for ex vivo expression of Foxp3 and IL-10. Each symbol represents pooled cTregs from 3-5 mice, horizontal lines show the mean and error bars the SD. Four independent experiments were performed. A Kruskal-Wallis test with a Dunn's post hoc test was performed in panels B, C, D, *** P <0.0001, ** P <0.01, * P <0.05. A Mann-Whitney U test was performed for experiments in panel E and _P_-values are shown where significant.
Fig 3. Ffar2 mediates SCFA effects on cTregs
(A) Tregs were isolated from the colon, small intestine, spleen and MLN of SPF and GF BALB/c mice, purified as in 1B and Ffar2 expression examined by qPCR. Each symbol represents data from 3-5 individual mice, horizontal lines show the mean and error bars the SD. Data reflect 3-7 independent experiments. (B) Lymphocytes were isolated from the colon, small intestine, spleen, and MLN of SPF Ffar2−/− and littermate Ffar2+/+ mice. Cells were stained for CD4, Foxp3, and Ffar2. Left panel depicts a representative flow cytometry histogram comparing colonic Ffar2 expression in Ffar2−/− vs littermate Ffar2+/+ mice. Right panel shows the MFI for Ffar2 for Tregs from the indicated sites. Bars show the mean, error bars SD, and data reflect 4 independent experiments. (C) Colonic LP lymphocytes were isolated from _Ffar2−/−_and littermate Ffar2+/+ mice exposed to propionate (P) or water alone and stained for CD4 and Foxp3. Left panel: Representative dot plots with percentage of CD4+Foxp3+ within the CD45+CD3+ population. Right panel: Foxp3+ Treg number for the left panel. Each symbol represents data from individual mice, horizontal lines show the mean and error bars the SD. (D) Ffar2−/− and littermate Ffar2+/+ cTregs were co-cultured with splenic Teff cells in media with or without propionate for 96 hrs. Percent suppression (y-axis) and Treg:Teff ratios (x-axis). Symbols represent the mean of 3 independent experiments and error bars show the SD. (E) cTregs were isolated from the LP of Ffar2−/− and littermate Ffar2+/+ mice, purified as in 1B, cultured in the presence of 0.1 mM propionate or media (pH and sodium matched) for 24hrs and examined for expression of HDAC 1,2,6,7 and 9 by RTqPCR. Bars show the mean and error bars the SD of 3 independent experiments (F) Whole cell extracts were generated from cTregs isolated from the LP of Ffar2−/− and littermate Ffar2+/+ mice, purified as in 1B, and cultured in the presence of 0.1 mM propionate or media (pH and sodium matched) for 24hrs. Samples were analyzed by western blotting for histone acetylation by examining levels of acetylated histone (H3K9), total histone levels were used as a loading control. The western blot shown is representative of two independent experiments with cTregs cell lysates pooled from 10-12 mice per group. A bar graph of densitometry ratios of acetylated Histone H3:total Histone H3 is shown. Bars represent the mean and error bars the SD. A Kruskal-Wallis test with a Dunn's post hoc test was performed for panels A and D, *** P < 0.001. The Mann-Whitney U test was performed for panels C and E. The student's t-test was performed for panel B and F.
Fig. 4. SCFA exposure ameliorates T cell transfer colitis in a Treg-intrinsic, _Ffar2_-dependent manner
BALB/c _Rag2_−/− mice were injected with CD4+CD45RBhiCD25lo naive T cells alone or in combination with Tregs. Following injection, mice received propionate, SCFA mix, or pH and sodium-matched drinking water. (A) Weekly percentage body weight change is shown across the experimental groups from experimental d0-d42. Symbols show the mean and error bars the SD. Data reflect three independent experiments. Colonic LP lymphocytes were isolated and stained for CD4 and Foxp3 and (B) percentage and (C) number of CD4+Foxp3+ within the CD45+CD3+ population are shown. Symbols represent data from individual mice, horizontal lines show the mean and error bars the SD. (D-F) C57BL/6 _Rag2_−/− mice were injected with CD4+CD45RBhiCD25lo naive T cells alone or in combination with Ffar2+/+ or Ffar2−/− Tregs. Following injection mice received propionate or pH and sodium-matched drinking water. (D) Histologic colitis score is shown along the y-axis, the treatment group and experimental conditions are shown along the x-axis. Colonic LP lymphocytes were isolated and (E) percentage and (F) number of CD4+Foxp3+ within the CD45+CD3+ population are shown. Symbols represent data from individual mice. Horizontal lines show the mean and error bars the SD. Panels D-F represent data from 2 independent experiments. The Kruskal-Wallis test with a Dunn's post hoc test was performed for panels A-F. ** P <0.01, * P <0.05.
Comment in
- Immunology. Feed your Tregs more fiber.
Bollrath J, Powrie F. Bollrath J, et al. Science. 2013 Aug 2;341(6145):463-4. doi: 10.1126/science.1242674. Science. 2013. PMID: 23908210 No abstract available. - Metabolites from intestinal microbes shape Treg.
Geuking MB, McCoy KD, Macpherson AJ. Geuking MB, et al. Cell Res. 2013 Dec;23(12):1339-40. doi: 10.1038/cr.2013.125. Epub 2013 Sep 10. Cell Res. 2013. PMID: 24018374 Free PMC article. - Regulatory T cells: distilling regulatory T cell inducers.
Papatriantafyllou M. Papatriantafyllou M. Nat Rev Immunol. 2013 Aug;13(8):546. doi: 10.1038/nri3506. Nat Rev Immunol. 2013. PMID: 24046840 No abstract available.
Similar articles
- Metabolites from intestinal microbes shape Treg.
Geuking MB, McCoy KD, Macpherson AJ. Geuking MB, et al. Cell Res. 2013 Dec;23(12):1339-40. doi: 10.1038/cr.2013.125. Epub 2013 Sep 10. Cell Res. 2013. PMID: 24018374 Free PMC article. - Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells.
Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Furusawa Y, et al. Nature. 2013 Dec 19;504(7480):446-50. doi: 10.1038/nature12721. Epub 2013 Nov 13. Nature. 2013. PMID: 24226770 - _Clostridium sporogenes_-derived metabolites protect mice against colonic inflammation.
Krause FF, Mangold KI, Ruppert AL, Leister H, Hellhund-Zingel A, Lopez Krol A, Pesek J, Watzer B, Winterberg S, Raifer H, Binder K, Kinscherf R, Walker A, Nockher WA, Taudte RV, Bertrams W, Schmeck B, Kühl AA, Siegmund B, Romero R, Luu M, Göttig S, Bekeredjian-Ding I, Steinhoff U, Schütz B, Visekruna A. Krause FF, et al. Gut Microbes. 2024 Jan-Dec;16(1):2412669. doi: 10.1080/19490976.2024.2412669. Epub 2024 Oct 14. Gut Microbes. 2024. PMID: 39397690 Free PMC article. - Induction of Treg cells in the mouse colonic mucosa: a central mechanism to maintain host-microbiota homeostasis.
Tanoue T, Honda K. Tanoue T, et al. Semin Immunol. 2012 Feb;24(1):50-7. doi: 10.1016/j.smim.2011.11.009. Epub 2011 Dec 14. Semin Immunol. 2012. PMID: 22172550 Review. - Modulation of microbially derived short-chain fatty acids on intestinal homeostasis, metabolism, and neuropsychiatric disorder.
Xiao S, Jiang S, Qian D, Duan J. Xiao S, et al. Appl Microbiol Biotechnol. 2020 Jan;104(2):589-601. doi: 10.1007/s00253-019-10312-4. Epub 2019 Dec 21. Appl Microbiol Biotechnol. 2020. PMID: 31865438 Review.
Cited by
- The microbiota-gut-brain axis: a potential target in the small-molecule compounds and gene therapeutic strategies for Parkinson's disease.
Jiao F, Zhou L, Wu Z. Jiao F, et al. Neurol Sci. 2024 Nov 15. doi: 10.1007/s10072-024-07878-x. Online ahead of print. Neurol Sci. 2024. PMID: 39546084 Review. - Added sugars and risk of osteoarthritis in adults: A case-control study based on National Health and Nutrition Examination Survey 2007-2018.
Liao X, Chen X, Zhou Y, Xing L, Shi Y, Huang G. Liao X, et al. PLoS One. 2024 Nov 14;19(11):e0313754. doi: 10.1371/journal.pone.0313754. eCollection 2024. PLoS One. 2024. PMID: 39541365 Free PMC article. - Insights into the unique roles of extracellular vesicles for gut health modulation: Mechanisms, challenges, and perspectives.
Wu Q, Kan J, Fu C, Liu X, Cui Z, Wang S, Le Y, Li Z, Liu Q, Zhang Y, Du J. Wu Q, et al. Curr Res Microb Sci. 2024 Oct 21;7:100301. doi: 10.1016/j.crmicr.2024.100301. eCollection 2024. Curr Res Microb Sci. 2024. PMID: 39525958 Free PMC article. Review. - IL-33 Induces a Switch in Intestinal Metabolites Revealing the Tryptophan Pathway as a Target for Inducing Allograft Survival.
Pinto C, Carrasco-Loncharic T, González-Mienert E, de Solminihac J, Gálvez-Jirón F, Cifuentes F, Pino-Lagos K. Pinto C, et al. Nutrients. 2024 Oct 27;16(21):3655. doi: 10.3390/nu16213655. Nutrients. 2024. PMID: 39519488 Free PMC article. - Cellular and Molecular Pathophysiology of Gestational Diabetes.
Torres-Torres J, Monroy-Muñoz IE, Perez-Duran J, Solis-Paredes JM, Camacho-Martinez ZA, Baca D, Espino-Y-Sosa S, Martinez-Portilla R, Rojas-Zepeda L, Borboa-Olivares H, Reyes-Muñoz E. Torres-Torres J, et al. Int J Mol Sci. 2024 Oct 30;25(21):11641. doi: 10.3390/ijms252111641. Int J Mol Sci. 2024. PMID: 39519193 Free PMC article. Review.
References
- Atarashi K, Umesaki Y, Honda K. Microbiotal influence on T cell subset development. Semin. Immunol. 2011;23:146–153. - PubMed
- Geuking MB, et al. Intestinal bacterial colonization of germ-free mice induces mutualistic regulatory T cell responses. Immunity. 34:794–806. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01CA154426/CA/NCI NIH HHS/United States
- F32 DK095506/DK/NIDDK NIH HHS/United States
- K08AI078942/AI/NIAID NIH HHS/United States
- F32DK095506/DK/NIDDK NIH HHS/United States
- R01 CA154426/CA/NCI NIH HHS/United States
- F32DK098826/DK/NIDDK NIH HHS/United States
- R01 GM099537/GM/NIGMS NIH HHS/United States
- F32 DK098826/DK/NIDDK NIH HHS/United States
- K08 AI078942/AI/NIAID NIH HHS/United States
- P30 DK034854/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases