Deciphering the rules by which 5'-UTR sequences affect protein expression in yeast - PubMed (original) (raw)
Deciphering the rules by which 5'-UTR sequences affect protein expression in yeast
Shlomi Dvir et al. Proc Natl Acad Sci U S A. 2013.
Erratum in
- Correction for Dvir et al., Deciphering the rules by which 5'-UTR sequences affect protein expression in yeast.
[No authors listed] [No authors listed] Proc Natl Acad Sci U S A. 2019 Jul 2;116(27):13701. doi: 10.1073/pnas.1909441116. Epub 2019 Jun 17. Proc Natl Acad Sci U S A. 2019. PMID: 31209029 Free PMC article. No abstract available.
Abstract
The 5'-untranslated region (5'-UTR) of mRNAs contains elements that affect expression, yet the rules by which these regions exert their effect are poorly understood. Here, we studied the impact of 5'-UTR sequences on protein levels in yeast, by constructing a large-scale library of mutants that differ only in the 10 bp preceding the translational start site of a fluorescent reporter. Using a high-throughput sequencing strategy, we obtained highly accurate measurements of protein abundance for over 2,000 unique sequence variants. The resulting pool spanned an approximately sevenfold range of protein levels, demonstrating the powerful consequences of sequence manipulations of even 1-10 nucleotides immediately upstream of the start codon. We devised computational models that predicted over 70% of the measured expression variability in held-out sequence variants. Notably, a combined model of the most prominent features successfully explained protein abundance in an additional, independently constructed library, whose nucleotide composition differed greatly from the library used to parameterize the model. Our analysis reveals the dominant contribution of the start codon context at positions -3 to -1, mRNA secondary structure, and out-of-frame upstream AUGs (uAUGs) to phenotypic diversity, thereby advancing our understanding of how protein levels are modulated by 5'-UTR sequences, and paving the way toward predictably tuning protein expression through manipulations of 5'-UTRs.
Keywords: AUG sequence context; computational prediction; mRNA folding; post-transcriptional regulation; upstream start codons.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
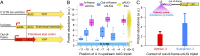
Accurate quantification of protein abundance in thousands of 5′-UTR sequence variants. (A) Schematics of the construction process of a 5′-UTR mutant library (see Materials and Methods for details). (B) Flow diagram of our experimental framework for measuring protein abundance levels. (1) We sorted our 5′-UTR mutant library into 24 bins according to the ratio between YFP and mCherry fluorescence (YFP/mCherry). (2) We PCR amplified each bin using bin-specific primers, each containing a unique 5-bp bar code sequence. We then pooled the obtained PCR products, subjected them to parallel DNA sequencing, and mapped each sequence read to a specific variant and to a specific bin (using the bin-specific bar codes). We quantified the mean protein abundance of each variant by using the distribution of its sequencing reads across expression bins (Materials and Methods). (3) As a control, we isolated individual strains from each bin, Sanger-sequenced the target 5′-UTR region, and measured the ratio between YFP and mCherry fluorescence by flow cytometry. (C) Our system provides highly accurate quantification of protein abundance levels. Shown is a comparison of the mean YFP-to-mCherry ratio as measured by flow cytometry (y axis; isolated variants), against abundance measurements estimated from our parallel sequencing approach (x axis; pooled variants) (R_2 = 0.98). (D) The figure depicts the distribution of protein abundance levels among 2,041 sequence variants. Note that mutations in the 5′-UTR (between positions −10 and −1; aaaacaaNNNNNNNNNN_AUG) generate approximately a sevenfold difference in protein levels.
Fig. 2.
The effect of specific nucleotides on protein levels. (A) The first three positions upstream of the start codon are key determinates of protein expression in our library. Shown are position-specific, logo-like representations of enrichment (over representation) and depletion (under representation) for highly (Left Upper) and lowly (Right Upper) expressed variants and high ribosome density genes in yeast (Lower Left). Over representation and under representation were calculated using the formula: _E_set[i,_k_] = _P_set[i,_k_]*log2(_P_set[i,_k_]/_P_back[i,_k_]), where, _P_set[i,_k_] denotes the probability of nucleotide i at position k in a subset of sequence variants (e.g., 10% of sequences with highest expression) and _P_back[i,_k_] is the probability of the same nucleotide at the same position in the appropriate background model (the remaining set of sequences). The relative height of individual symbols (A, C, G, or T) equals to _E_set[i,_k_], whereas enrichment and depletion are indicated by positive and negative _E_set[i,_k_] values, respectively. Statistical significance was quantified by using a two-tailed Fisher's exact test, controlling for a false discovery rate (74) of 1%. Nucleotides with false discovery rate-corrected P values greater or equal to 0.01 are colored gray. The genome-wide analysis is based on average ribosome densities of two biological replicates (5). (B) Purine at position −3 supports high levels of expression. Box plots of protein abundance (y axis) in the presence (orange; n = 1,164) and absence (magenta; n = 877) of a −3 purine. Outliers (gray plus signs) are defined as expression levels that are either larger than the upper quartile or smaller than the lower quartile by more than 1.5 times the interquartile range. See
SI Appendix, Fig. S9
, for position-specific comparisons of protein levels in the presence and absence of each of the four nucleotide bases.
Fig. 3.
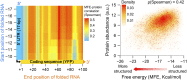
Stable mRNA secondary structures are correlated with reduced protein levels. The left panel shows a heat map of Spearman correlations, where each correlation denotes the association between the minimum free energy (MFE) of a given mRNA region and protein levels, in 2,041 variants. We used the region between −10 and −1 as the smallest folding segment, while moving upstream and/or downstream in 1-bp steps (up to −17 and +100). The y and x axes represent the start (in 5′-UTR) and end (in coding region) position of each folded segment, respectively. For each correlation, we obtained a two-sided P value by performing 100,000 permutation tests of shuffled expression values. For clarity of the figure, we removed folding segments that include only 5′-UTR nucleotides, without the YFP coding region (the Spearman's ρ for these regions is 0.09 ± 0.003; median ± MAD; P < 0.001). The right panel shows an example of the above relationship for the mRNA segment between positions −15 to +50 (Spearman's ρ = 0.42, P < 10−86). We obtained equivalent results, albeit with slightly higher correlations, by using the ensemble free energy measure (
SI Appendix, Fig. S10
, and related
SI Appendix, Note S4
). Folding free energies were computed with RNAfold from the Vienna RNA package at a folding temperature of 30 °C (51).
Fig. 4.
Out-of-frame upstream start codons attenuate protein expression. (A) Schematic representation of in-frame and out-of-frame uAUGs. We note that our definition of a uAUG (see text) differs from the traditional upstream ORF (uORF) which usually contains a uAUG triplet and an in-frame stop codon, fully embedded within the 5′-UTR. (B) Out-of-frame uAUGs decrease protein levels. Shown are position-specific box plots of protein abundance (y axis) for out-of-frame uAUGs (light blue), in-frame uAUGs (magenta), and uAUG-free sequences (yellow). The x axis indicates the position of the A nucleotide in the uAUG triplet (relative to the main ORF). Note that RPL8A contains an adenine nucleotide at position −11 (there are no mutations at this position) and hence may form a uAUG codon. P values (see text) were obtained using two-sided Wilcoxon rank sum and Kolmogorov–Smirnov tests. (C) The inhibitory effect of out-of-frame uAUGs is augmented by optimal uAUG context. The graph represents a comparison of protein abundance levels of out-of-frame uAUG variants with optimal (purine, red bar) and suboptimal (pyrimidine, light blue bar) nucleotides at position −3 upstream of the uAUG codon (two-sided Wilcoxon rank sum, P < 0.01). Error bars represent the SD.
Fig. 5.
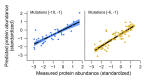
Quantitative models predict over 70% of the variation in protein levels in held-out test sets. (A) Prediction performance reaches a plateau at 20–25 predictors. The colored lines depict the average _R_2 (y axis) obtained in a 10-fold cross-validation (CV) strategy for linear models that each uses a different number of feature predictors (x axis), for both the train (magenta) and test (orange) sets. The colored areas above and below the lines represent the SD. Note that a high fraction of the expression variation can be explained, for example, by using 20 predictors (mean ± SD, test _R_2 = 0.69 ± 0.05), whereas adding further features only marginally improves performance (mean ± SD, test _R_2 = 0.74 ± 0.05; 40 predictors). (B) A combined model with as few as 13 predictors explains 68% of the expression variation in our library. A comparison between measured protein abundance (x axis) and abundance levels predicted by our combined model (y axis), using the 13 most informative predictors. Protein abundance levels are expressed as standardized scores (during model construction, continuous predictors and measured protein levels were standardized to zero mean and unit SD; binary predictors were coded as 1 and 0). (C) A graphical depiction of our combined model. Shown are the 13 predictors (y axis) included in our combined model with their respective model coefficients (x axis): (i) Nucleotide preferences at positions −3 to −1 (six features, orange); (ii) mRNA secondary structure (abbreviated as RSS, light blue); (iii) out-of-frame uAUGs (pink); and (iv) short uncharacterized _k_-mers (five features, red). Positive and negative coefficients are marked in red and light blue dots, respectively. Numbers in brackets indicate the quantity of variants associated with each predictor. (D) Predictive power of different feature groups. We constructed a linear regression model for each of the four feature groups from C, reporting the proportion of variance explained (_R_2) by each group. In addition, we found that the top six most dominant predictors in our combined model account for 61% of the expression variation (dark orange, Bottom). The six predictors are as follows: a purine at position −3, adenine at position −1, mRNA secondary structure, out-of-frame uAUGs, GG-dinucleotides, and a CACC pattern. As in B, protein abundance levels are expressed as standardized scores. _R_2 values indicate the average performance obtained on test sets, using 10-fold CV.
Fig. 6.
Our combined model successfully accounts for most of the expression variation in two additional, independently constructed RPL8A 5′-UTR mutant pools. Shown is a comparison between measured protein abundance (x axes) and predictions made by our combined model (y axes) for: (i) An A/C-rich collection with random mutations between positions −10 to −1 (44 variants; Left; _R_2 = 0.69), and (ii) a G/C-rich collection with random perturbations between positions −6 to −1 (65 variants; Right; _R_2 = 0.71). The predictions were based on standardized features (z scores), whereby we used the mean and SD obtained for a specific feature in the large-scale library to standardize the same feature in the new collections (assuming that all pools come from the same probability distribution). Binary predictors were coded as 1 and 0. Linear fits are depicted in black with their respective 95% confidence intervals (the colored area above and below the lines).
Similar articles
- Small, synthetic, GC-rich mRNA stem-loop modules 5' proximal to the AUG start-codon predictably tune gene expression in yeast.
Lamping E, Niimi M, Cannon RD. Lamping E, et al. Microb Cell Fact. 2013 Jul 29;12:74. doi: 10.1186/1475-2859-12-74. Microb Cell Fact. 2013. PMID: 23895661 Free PMC article. - Presence of ATG triplets in 5' untranslated regions of eukaryotic cDNAs correlates with a 'weak' context of the start codon.
Rogozin IB, Kochetov AV, Kondrashov FA, Koonin EV, Milanesi L. Rogozin IB, et al. Bioinformatics. 2001 Oct;17(10):890-900. doi: 10.1093/bioinformatics/17.10.890. Bioinformatics. 2001. PMID: 11673233 - mRNA sequences influencing translation and the selection of AUG initiator codons in the yeast Saccharomyces cerevisiae.
Yun DF, Laz TM, Clements JM, Sherman F. Yun DF, et al. Mol Microbiol. 1996 Mar;19(6):1225-39. doi: 10.1111/j.1365-2958.1996.tb02468.x. Mol Microbiol. 1996. PMID: 8730865 - A helicase links upstream ORFs and RNA structure.
Jankowsky E, Guenther UP. Jankowsky E, et al. Curr Genet. 2019 Apr;65(2):453-456. doi: 10.1007/s00294-018-0911-z. Epub 2018 Nov 27. Curr Genet. 2019. PMID: 30483885 Free PMC article. Review.
Cited by
- Controlling gene expression with deep generative design of regulatory DNA.
Zrimec J, Fu X, Muhammad AS, Skrekas C, Jauniskis V, Speicher NK, Börlin CS, Verendel V, Chehreghani MH, Dubhashi D, Siewers V, David F, Nielsen J, Zelezniak A. Zrimec J, et al. Nat Commun. 2022 Aug 30;13(1):5099. doi: 10.1038/s41467-022-32818-8. Nat Commun. 2022. PMID: 36042233 Free PMC article. - Functional analysis of the three HMA4 copies of the metal hyperaccumulator Arabidopsis halleri.
Nouet C, Charlier JB, Carnol M, Bosman B, Farnir F, Motte P, Hanikenne M. Nouet C, et al. J Exp Bot. 2015 Sep;66(19):5783-95. doi: 10.1093/jxb/erv280. Epub 2015 Jun 4. J Exp Bot. 2015. PMID: 26044091 Free PMC article. - Zinc triggers a complex transcriptional and post-transcriptional regulation of the metal homeostasis gene FRD3 in Arabidopsis relatives.
Charlier JB, Polese C, Nouet C, Carnol M, Bosman B, Krämer U, Motte P, Hanikenne M. Charlier JB, et al. J Exp Bot. 2015 Jul;66(13):3865-78. doi: 10.1093/jxb/erv188. Epub 2015 Apr 21. J Exp Bot. 2015. PMID: 25900619 Free PMC article. - The relationship of mRNA with protein expression in CD8+ T cells associates with gene class and gene characteristics.
Nicolet BP, Wolkers MC. Nicolet BP, et al. PLoS One. 2022 Oct 19;17(10):e0276294. doi: 10.1371/journal.pone.0276294. eCollection 2022. PLoS One. 2022. PMID: 36260607 Free PMC article. - Regulation of ribosomal protein genes: An ordered anarchy.
Petibon C, Malik Ghulam M, Catala M, Abou Elela S. Petibon C, et al. Wiley Interdiscip Rev RNA. 2021 May;12(3):e1632. doi: 10.1002/wrna.1632. Epub 2020 Oct 10. Wiley Interdiscip Rev RNA. 2021. PMID: 33038057 Free PMC article. Review.
References
- Raveh-Sadka T, et al. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat Genet. 2012;44(7):743–750. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases