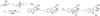Radical SAM-mediated methylation reactions - PubMed (original) (raw)
Review
Radical SAM-mediated methylation reactions
Danica Galonić Fujimori. Curr Opin Chem Biol. 2013 Aug.
Abstract
A subset of enzymes that belong to the radical S-adenosylmethionine (SAM) superfamily is able to catalyze methylation reactions. Substrates of these enzymes are distinct from the nucleophilic substrates that undergo methylation by a polar mechanism. Recently, activities of several radical SAM methylating enzymes have been reconstituted in vitro and their mechanisms of catalysis investigated. The RNA modifying enzymes RlmN and Cfr catalyze methylation via a methyl synthase mechanism. These enzymes use SAM in two distinct roles: as a source of a methyl group transferred to a conserved cysteine and as a source of 5'-deoxyadenosyl radical (5'-dA). Hydrogen atom abstraction by this species generates a thiomethylene radical which adds into the RNA substrate, forming an enzyme-substrate covalent adduct. In another recent study, methylation of the indole moiety of tryptophan by the radical SAM and cobalamin-binding domain enzyme TsrM has been reconstituted. Methylcobalamin serves as an intermediate methyl donor in TsrM, and is proposed to transfer the methyl group as a methyl radical. Interestingly, despite the presence of the radical SAM motif, no reductive cleavage of SAM has been observed in this methylation. These important reconstitutions set the stage for further studies on mechanisms of radical methylation.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
Figure 1
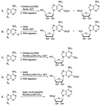
Selected molecules that contain methyl groups or their derivatives introduced by radical SAM methylating enzymes. Groups introduced by these enzymes are shown in red. Polytheonamides A and B differ only in the absolute configuration of the sulfoxide [58].
Figure 2
Summary of labeling experiments used to elucidate the mechanism of methylation by radical SAM methyl synthases RlmN [12,42]. Experiments in panels A and B were performed under multiple turnover conditions, whereas experiments in panels C–E were performed under single turnover conditions. SDT = sodium dithionite. Flv = flavodoxin. Flx = flavodoxin reductase. R1 = nucleotides 2447–2778, 23S rRNA. R2 = nucleotides 2500–2506, 23S rRNA. R3 = nucleotides 2018–2788, 23S rRNA. The RlmN substrate adenosine is in position 2503 in all pictured RNA constructs.
Figure 3
Proposed mechanism of RNA methylation by RlmN [12]. The covalent intermediate, trapped by mutagenesis of Cys118 to Ala [13], is shown in the inset. SAM = _S_-adenosyl-L-methionine. SAH = _S_-adenosyl-L-homocysteine. 5′-dA• = 5′-deoxyadenosyl radical.
Figure 4
Proposed mechanism of the C2 methylation of the indole moiety of tryptophan by TsrM [30]. The Trp substrate is putatively coordinated by the iron-sulfur cluster. Reduction of Cob(II)alamin by the reduced [4Fe-4S] cluster may result in transient formation of the nucleophilic Co(I), which may react with SAM to regenerate methylcobalamin cofactor. Following addition of the methyl radical, oxidation of the Trp radical might be achieved by the oxidized cluster. SAM = _S_-adenosyl-L-methionine. SAH = _S_-adenosyl-L-homocysteine.
Similar articles
- Radical-mediated enzymatic methylation: a tale of two SAMS.
Zhang Q, van der Donk WA, Liu W. Zhang Q, et al. Acc Chem Res. 2012 Apr 17;45(4):555-64. doi: 10.1021/ar200202c. Epub 2011 Nov 18. Acc Chem Res. 2012. PMID: 22097883 Free PMC article. Review. - Efficient methylation of C2 in l-tryptophan by the cobalamin-dependent radical _S_-adenosylmethionine methylase TsrM requires an unmodified N1 amine.
Blaszczyk AJ, Wang B, Silakov A, Ho JV, Booker SJ. Blaszczyk AJ, et al. J Biol Chem. 2017 Sep 15;292(37):15456-15467. doi: 10.1074/jbc.M117.778548. Epub 2017 Jul 26. J Biol Chem. 2017. PMID: 28747433 Free PMC article. - Understanding the role of electron donors in the reaction catalyzed by Tsrm, a cobalamin-dependent radical S-adenosylmethionine methylase.
Blaszczyk AJ, Knox HL, Booker SJ. Blaszczyk AJ, et al. J Biol Inorg Chem. 2019 Sep;24(6):831-839. doi: 10.1007/s00775-019-01689-8. Epub 2019 Jul 26. J Biol Inorg Chem. 2019. PMID: 31350635 Free PMC article. - Structural basis for non-radical catalysis by TsrM, a radical SAM methylase.
Knox HL, Chen PY, Blaszczyk AJ, Mukherjee A, Grove TL, Schwalm EL, Wang B, Drennan CL, Booker SJ. Knox HL, et al. Nat Chem Biol. 2021 Apr;17(4):485-491. doi: 10.1038/s41589-020-00717-y. Epub 2021 Jan 18. Nat Chem Biol. 2021. PMID: 33462497 Free PMC article. - Mechanistic diversity of radical S-adenosylmethionine (SAM)-dependent methylation.
Bauerle MR, Schwalm EL, Booker SJ. Bauerle MR, et al. J Biol Chem. 2015 Feb 13;290(7):3995-4002. doi: 10.1074/jbc.R114.607044. Epub 2014 Dec 4. J Biol Chem. 2015. PMID: 25477520 Free PMC article. Review.
Cited by
- Methylated nucleosides in tRNA and tRNA methyltransferases.
Hori H. Hori H. Front Genet. 2014 May 23;5:144. doi: 10.3389/fgene.2014.00144. eCollection 2014. Front Genet. 2014. PMID: 24904644 Free PMC article. Review. - Mechanism of a Class C Radical S-Adenosyl-l-methionine Thiazole Methyl Transferase.
Zhang Z, Mahanta N, Hudson GA, Mitchell DA, van der Donk WA. Zhang Z, et al. J Am Chem Soc. 2017 Dec 27;139(51):18623-18631. doi: 10.1021/jacs.7b10203. Epub 2017 Dec 15. J Am Chem Soc. 2017. PMID: 29190095 Free PMC article. - Does Viperin Function as a Radical S-Adenosyl-l-methionine-dependent Enzyme in Regulating Farnesylpyrophosphate Synthase Expression and Activity?
Makins C, Ghosh S, Román-Meléndez GD, Malec PA, Kennedy RT, Marsh EN. Makins C, et al. J Biol Chem. 2016 Dec 23;291(52):26806-26815. doi: 10.1074/jbc.M116.751040. Epub 2016 Nov 10. J Biol Chem. 2016. PMID: 27834682 Free PMC article. - Replacement of the Cobalt Center of Vitamin B12 by Nickel: Nibalamin and Nibyric Acid Prepared from Metal-Free B12 Ligands Hydrogenobalamin and Hydrogenobyric Acid.
Kieninger C, Wurst K, Podewitz M, Stanley M, Deery E, Lawrence AD, Liedl KR, Warren MJ, Kräutler B. Kieninger C, et al. Angew Chem Int Ed Engl. 2020 Nov 2;59(45):20129-20136. doi: 10.1002/anie.202008407. Epub 2020 Sep 2. Angew Chem Int Ed Engl. 2020. PMID: 32686888 Free PMC article. - The Hydrogenobyric Acid Structure Reveals the Corrin Ligand as an Entatic State Module Empowering B12 Cofactors for Catalysis.
Kieninger C, Deery E, Lawrence AD, Podewitz M, Wurst K, Nemoto-Smith E, Widner FJ, Baker JA, Jockusch S, Kreutz CR, Liedl KR, Gruber K, Warren MJ, Kräutler B. Kieninger C, et al. Angew Chem Int Ed Engl. 2019 Jul 29;58(31):10756-10760. doi: 10.1002/anie.201904713. Epub 2019 Jun 26. Angew Chem Int Ed Engl. 2019. PMID: 31115943 Free PMC article.
References
- Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. - PMC - PubMed
- Frey PA, Magnusson OT. S-Adenosylmethionine: a wolf in sheep's clothing, or a rich man's adenosylcobalamin? Chem Rev. 2003;103:2129–2148. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources