Ancient DNA analysis of mid-holocene individuals from the Northwest Coast of North America reveals different evolutionary paths for mitogenomes - PubMed (original) (raw)
. 2013 Jul 3;8(7):e66948.
doi: 10.1371/journal.pone.0066948. Print 2013.
John Lindo, Cris E Hughes, Jesse W Johnson, Alvaro G Hernandez, Brian M Kemp, Jian Ma, Ryan Cunningham, Barbara Petzelt, Joycellyn Mitchell, David Archer, Jerome S Cybulski, Ripan S Malhi
Affiliations
- PMID: 23843972
- PMCID: PMC3700925
- DOI: 10.1371/journal.pone.0066948
Ancient DNA analysis of mid-holocene individuals from the Northwest Coast of North America reveals different evolutionary paths for mitogenomes
Yinqiu Cui et al. PLoS One. 2013.
Abstract
To gain a better understanding of North American population history, complete mitochondrial genomes (mitogenomes) were generated from four ancient and three living individuals of the northern Northwest Coast of North America, specifically the north coast of British Columbia, Canada, current home to the indigenous Tsimshian, Haida, and Nisga'a. The mitogenomes of all individuals were previously unknown and assigned to new sub-haplogroup designations D4h3a7, A2ag and A2ah. The analysis of mitogenomes allows for more detailed analyses of presumed ancestor-descendant relationships than sequencing only the HVSI region of the mitochondrial genome, a more traditional approach in local population studies. The results of this study provide contrasting examples of the evolution of Native American mitogenomes. Those belonging to sub-haplogroups A2ag and A2ah exhibit temporal continuity in this region for 5000 years up until the present day. Of possible associative significance is that archaeologically identified house structures in this region maintain similar characteristics for this same period of time, demonstrating cultural continuity in residence patterns. The individual dated to 6000 years before present (BP) exhibited a mitogenome belonging to sub-haplogroup D4h3a. This sub-haplogroup was earlier identified in the same general area at 10300 years BP on Prince of Wales Island, Alaska, and may have gone extinct, as it has not been observed in any living individuals of the Northwest Coast. The presented case studies demonstrate the different evolutionary paths of mitogenomes over time on the Northwest Coast.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
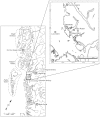
Figure 1. Map of study area.
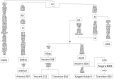
Figure 2. Phylogeny of complete mitochondrial genomes sequenced in this study.
Mutations are transitions unless specified. Transversions are indicated by an A, G, C, or T after the nucleotide position. Insertions are indicated by an “i”, deletions are indicated by a “d”, recurrent mutations are underlined, and mutations back to the rCRS nucleotide are designated by a “@”. The C stretch length polymorphism in region 303–315 was disregarded in the tree. The sample “Haida 9″ was analyzed in Schurr et al. (2012). All other samples were analyzed in this study.
Similar articles
- Ancient individuals from the North American Northwest Coast reveal 10,000 years of regional genetic continuity.
Lindo J, Achilli A, Perego UA, Archer D, Valdiosera C, Petzelt B, Mitchell J, Worl R, Dixon EJ, Fifield TE, Rasmussen M, Willerslev E, Cybulski JS, Kemp BM, DeGiorgio M, Malhi RS. Lindo J, et al. Proc Natl Acad Sci U S A. 2017 Apr 18;114(16):4093-4098. doi: 10.1073/pnas.1620410114. Epub 2017 Apr 4. Proc Natl Acad Sci U S A. 2017. PMID: 28377518 Free PMC article. - Patterns of mtDNA diversity in northwestern North America.
Malhi RS, Breece KE, Shook BA, Kaestle FA, Chatters JC, Hackenberger S, Smith DG. Malhi RS, et al. Hum Biol. 2004 Feb;76(1):33-54. doi: 10.1353/hub.2004.0023. Hum Biol. 2004. PMID: 15222679 Review. - Ancient mitogenomes from the Southern Pampas of Argentina reflect local differentiation and limited extra-regional linkages after rapid initial colonization.
Motti JMB, Pauro M, Scabuzzo C, García A, Aldazábal V, Vecchi R, Bayón C, Pastor N, Demarchi DA, Bravi CM, Reich D, Cabana GS, Nores R. Motti JMB, et al. Am J Biol Anthropol. 2023 Jun;181(2):216-230. doi: 10.1002/ajpa.24727. Epub 2023 Mar 15. Am J Biol Anthropol. 2023. PMID: 36919783 - A South American Prehistoric Mitogenome: Context, Continuity, and the Origin of Haplogroup C1d.
Sans M, Figueiro G, Hughes CE, Lindo J, Hidalgo PC, Malhi RS. Sans M, et al. PLoS One. 2015 Oct 28;10(10):e0141808. doi: 10.1371/journal.pone.0141808. eCollection 2015. PLoS One. 2015. PMID: 26509686 Free PMC article. - Mitochondrial DNA and prehistoric settlements: native migrations on the western edge of North America.
Eshleman JA, Malhi RS, Johnson JR, Kaestle FA, Lorenz J, Smith DG. Eshleman JA, et al. Hum Biol. 2004 Feb;76(1):55-75. doi: 10.1353/hub.2004.0019. Hum Biol. 2004. PMID: 15222680 Review.
Cited by
- Unraveling the Genetic Threads of History: mtDNA HVS-I Analysis Reveals the Ancient Past of the Aburra Valley.
Uricoechea Patiño D, Collins A, Romero García OJ, Santos Vecino G, Aristizábal Espinosa P, Bernal Villegas JE, Benavides Benitez E, Vergara Muñoz S, Briceño Balcázar I. Uricoechea Patiño D, et al. Genes (Basel). 2023 Nov 2;14(11):2036. doi: 10.3390/genes14112036. Genes (Basel). 2023. PMID: 38002979 Free PMC article. - Unraveling the mitochondrial phylogenetic landscape of Thailand reveals complex admixture and demographic dynamics.
Jaisamut K, Pitiwararom R, Sukawutthiya P, Sathirapatya T, Noh H, Worrapitirungsi W, Vongpaisarnsin K. Jaisamut K, et al. Sci Rep. 2023 Nov 21;13(1):20396. doi: 10.1038/s41598-023-47762-w. Sci Rep. 2023. PMID: 37990137 Free PMC article. - A paleogenome from a Holocene individual supports genetic continuity in Southeast Alaska.
Aqil A, Gill S, Gokcumen O, Malhi RS, Reese EA, Smith JL, Heaton TT, Lindqvist C. Aqil A, et al. iScience. 2023 Apr 8;26(5):106581. doi: 10.1016/j.isci.2023.106581. eCollection 2023 May 19. iScience. 2023. PMID: 37138779 Free PMC article. - Species identification and mitochondrial genomes of ancient fish bones from the Riverine Kachemak tradition of the Kenai Peninsula, Alaska.
de Flamingh A, Mallott EK, Roca AL, Boraas AS, Malhi RS. de Flamingh A, et al. Mitochondrial DNA B Resour. 2018 Apr 1;3(1):409-411. doi: 10.1080/23802359.2018.1456371. Mitochondrial DNA B Resour. 2018. PMID: 33474186 Free PMC article. - Fostering Responsible Research on Ancient DNA.
Wagner JK, Colwell C, Claw KG, Stone AC, Bolnick DA, Hawks J, Brothers KB, Garrison NA. Wagner JK, et al. Am J Hum Genet. 2020 Aug 6;107(2):183-195. doi: 10.1016/j.ajhg.2020.06.017. Am J Hum Genet. 2020. PMID: 32763189 Free PMC article. Review.
References
- Perego UA, Achilli A, Angerhofer N, Accetturo M, Pala M, et al. (2009) Distinctive paleo-indian migration routes from beringia marked by two rare mtDNA haplogroups. Curr Biol 19: 1–8. - PubMed
Publication types
MeSH terms
Grants and funding
This study was funded by the National Science Foundation grant BCS-0745459 and BCS-1025139 to RSM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources