ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon - PubMed (original) (raw)
ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon
Jenish R Patel et al. EMBO Rep. 2013 Sep.
Abstract
The cytosolic pathogen sensor RIG-I is activated by RNAs with exposed 5'-triphosphate (5'-ppp) and terminal double-stranded structures, such as those that are generated during viral infection. RIG-I has been shown to translocate on dsRNA in an ATP-dependent manner. However, the precise role of the ATPase activity in RIG-I activation remains unclear. Using in vitro-transcribed Sendai virus defective interfering RNA as a model ligand, we show that RIG-I oligomerizes on 5'-ppp dsRNA in an ATP hydrolysis-dependent and dsRNA length-dependent manner, which correlates with the strength of type-I interferon (IFN-I) activation. These results establish a clear role for the ligand-induced ATPase activity of RIG-I in the stimulation of the IFN response.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
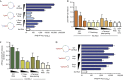
SeV DI RNA is a potent RIG-I-dependent inducer of IFN-I. (A) Graphic illustrates different RNAs produced during infection of SeV. SeV DI produced from the anti-genomic positive sense (+) RNA consists of both negative and positive sense sequences of the genomic and anti-genomic RNAs resulting in a copy-back structure. The right panel shows the 546-nt SeV DI RNA mapped to the genomic/anti-genomic sequence and a predicted structure of the RNA (RNAfold). Colours on the DI RNA representations indicate base-pair probability (as indicated, red=1; purple=0). (B) 293T-IFNβ-FF-Luc cells were transfected with 50 ng of indicated RNAs and a luciferase assay was performed 24 h later to measure IFNβ promoter-driven luciferase activity. (C) 293T-IFNβ-FF-Luc cells were transfected with control or RIG-I short interfering RNA and 24 h later transfected with 5 and 1 ng of IVT DI RNA and 200 and 40 ng poly (I:C), followed by luciferase assay 24 h later to measure IFNβ promoter-driven luciferase activity. (D) Lysates from 293T cells expressing FL RIG-I or truncated RIG-I Hel, RIG-I C-terminal RD domain or RIG-I Hel-RD were incubated with 0.25 μg of RNA, RIG-I–RNA complexes were immunoprecipitated, and RNA and protein fractions were isolated. RNA was transfected into 293T-IFNβ-FF-Luc cells and 24 h later IFNβ promoter-driven luciferase activity was measured by luciferase assay. Protein fractions were subjected to immunoblotting using HA antibody to assess pull-down efficiency. Data are representative of at least three independent experiments and error bars indicate mean±s.d. DI, defective interfering; FL, full length; Hel, Helicase domain; Hel-RD, Helicase regulatory domain; IFN-I, type-I interferon; IVT, in vitro transcribed; RD, regulatory domain; SeV, Sendai virus.
Figure 2
Exposed 5′-ppp and terminal dsRNA, but not loop structure, are important features of SeV DI RNA for RIG-I-mediated IFN-I activation. (A) 25 fmol and five-fold dilutions of WT, 5′ overhang and Δ terminal base-pairing RNAs were transfected or not into 293T-IFNβ-FF-Luc cells and 24 h later IFNβ promoter-driven luciferase activity was measured by a luciferase assay. (B) 500 fmol and three-fold dilutions of Biotin-UTP-labelled WT, 5′ overhang and Δ terminal base-pairing RNAs were immobilized onto NeutrAvidin-coated wells and incubated with lysates from HA-RIG-I-expressing 293T cells. The levels of bound RIG-I were determined by measuring the absorbance/HRP activity of HA-HRP antibody. (C) 0.5 μg of purified His-HA-RIG-I was incubated with 100 or 50 fmol of WT, 5′ overhang, Δ terminal base-pairing or no RNA in the presence of 0.5 mM ATP and 2.5 mM Mg2+ at 37 °C for 25 min. Released phosphates were measured using Malachite Green-based reagent at an absorbance of 620 nm. (D) 25 fmol and five-fold dilutions of WT, half-loop and short loop RNAs were transfected or not into 293T-IFNβ-FF-Luc cells and 24 h later IFNβ promoter-driven luciferase activity was measured by a luciferase assay. Data are representative of at least three independent experiments and error bars indicate mean±s.d. Log10 in Figs 2A and 2D refers to the scale on the x-axis. 5′-ppp, 5′-triphosphate; DI, defective interfering; dsRNA, double-stranded RNA; HRP, horseradish peroxidase; IFNβ, interferon β; SeV, Sendai virus; WT, wild type.
Figure 3
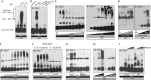
dsRNA length-dependent oligomerization is a mechanism for the high immunostimulatory activity of SeV DI RNA. (A) 5 ng and five-fold dilutions of WT, 46- and 25-bp stem RNAs were transfected or not into 293T-IFNβ-FF-Luc cells and 24 h later IFNβ promoter-driven luciferase activity was measured by a luciferase assay. (B) 500 fmol and three-fold dilutions of Biotin-UTP-labelled WT, 46- and 25-bp stem RNAs were immobilized onto NeutrAvidin-coated wells and incubated with lysates from HA-RIG-I expressing 293T cells. The levels of bound RIG-I were determined by measuring the absorbance/HRP activity of HA-HRP antibody. (C) 100 and 50 fmol of RNAs was incubated with 0.5 μg of RIG-I in the presence of 0.5 mM ATP and 2.5 mM Mg2+ at 37 °C for 25 min. Released phosphates were measured by a colorimetric ATPase assay at absorbance 620 nm. (D) Lysates from cells expressing eYFP-RIG-I or HA-RIG-I were mixed and incubated with 1.25 and 5 pmol of WT, 46- and 25-bp stem RNA or no RNA, eYFP-RIG-I was immunoprecipitated with GFP antibody and eYFP-RIG-I and HA-RIG-I levels were assessed by SDS–PAGE and immunoblotting with GFP or HA antibodies. Levels of input HA-RIG-I in WCL were determined by SDS–PAGE and immunoblotting with HA antibody. (E) 1 μg of RIG-I was incubated with 1.25 or 0.625 pmol of WT, 46- and 25-bp stem RNAs or no RNA in the presence of 0.5 mM ATP and 2.5 mM Mg2+ for 25 min at 37 °C and RIG-I complexes were analysed by NativePAGE and immunoblotting. Data are representative of at least three independent experiments and error bars indicate mean±s.d. Log10 in Fig 3A refers to the scale on the x-axis. DI, defective interfering; dsRNA, double-stranded RNA; GFP, green fluorescent protein; HRP, horseradish peroxidase; SeV, Sendai virus; WCL, whole cell lysates; WT, wild type.
Figure 4
ATP hydrolysis drives oligomerization of RIG-I on 5′-ppp dsRNA. (A,B) 0.2 μg of indicated IVT RNAs was incubated with 1 μg of RIG-I in the presence of 1 mM ATP and 2.5 mM Mg2+ for 25 min at 37 °C, analysed by NativePAGE and immunoblotted for RIG-I. (C) Left panel: 0.5 μg of RIG-I was incubated with 0.05, 0.075, 0.1, 0.175, 0.25, 0.5, 1 and 2 μg of RNA or no RNA in the presence of 1 mM ATP and 2.5 mM Mg2+ for 25 min at 37 °C and analysed. Right panel: 0.5 μg of RNA was incubated with 0.05, 0.1, 0.25, 0.5, 1, 2, 3, 4 or 5 μg of RIG-I in the presence of 1 mM ATP and 2.5 mM Mg2+ for 25 min at 37 °C and analysed. (D) 0.2 μg of Biotin-RNA was incubated with 0.05, 0.25, 0.5 and 1 μg of RIG-I in the presence of 1 mM ATP and 2.5 mM Mg2+ for 25 min at 37 °C, analysed by nativePAGE and immunoblotted for Biotin-RNA and RIG-I. (E) 2 μg of RIG-I was incubated with 0.3 μg of RNA in the absence or presence of 0.05, 0.2, 0.5, 1, 2, 3, 4 and 5 mM of ATP and 2.5 mM Mg2+ for 25 min at 37 °C and analysed. (F) 2 μg of RIG-I was incubated with 0.3 μg of RNA with or without 2 mM ATP for 5, 15, 25 and 45 min at 37 °C and RIG-I oligomerization was analysed. (G) 2 μg of RIG-I was incubated with 0.3 μg of RNA in the presence of 1, 0.5 and 0.25 mM of ATP or ADPCP for 25 min at 37 °C and native complexes were analysed. (H) 0.25 μg of RNA was incubated with 0.05, 0.3 and 1 μg of either WT or D372N RIG-I with 1 mM of ATP and 2.5 mM Mg2+ for 25 min at 37 °C and analysed. (I) 0.2 μg of RNA was incubated with 1 μg of RIG-I in the presence or absence of 1 mM ATP and 2.5 mM Mg2+ for 15 min at 37 °C, followed by addition of 0, 0.01 and 0.1 units of RNase V1 at RT for 15 min and RIG-I oligomerization was analysed. Data are representative of two to three independent experiments. 5′-ppp, 5′-triphosphate; dsRNA, double-stranded RNA; IVT, in vitro transcribed.
Figure 5
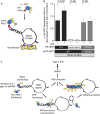
Mechanism for ATP-driven formation of RIG-I oligomers on RNA. (A) Schematic of the experiment highlighting immobilization of Biotin-labelled RNA onto NeutrAvidin and saturation with eYFP-RIG-I, followed by binding of HA-RIG-I in the presence or absence of ATP. (B) 80 ng of Biotin-labelled DI RNA was immobilized on NeutrAvidin-coated wells, washed and 1 μg of eYFP-RIG-I added (WT or D372N mutant). Following washes, 0.2 μg of HA-RIG-I (WT, K858A-K861A or D372N) was added in the presence or absence of 1 mM ATP as indicated. Bound HA-RIG-I was detected using α-HA-HRP antibody. Data are representative of three independent experiments and error bars indicate mean±s.d. The _P_-value was calculated using a Student’s unpaired t test. (C) A proposed model of RIG-I activation. Upon binding to 5′-ppp on dsRNA ligand, RIG-I is activated with a conformation change and ATP hydrolysis. Our data suggest that ATP hydrolysis by RNA-bound RIG-I allows exposure of 5′-ppp and recruitment of additional RIG-I molecules on long dsRNA forming RIG-I oligomers. As demonstrated previously, upon K63-linked polyubiquitination or polyubiquitin binding, RIG-I is further activated and binds MAVS to induce IFN-I production. 5′-ppp, 5′-triphosphate; DI, defective interfering; dsRNA, double-stranded RNA; HRP, horseradish peroxidase; IFN-I, type-1 interferon; MAVS, mitochondrial antiviral signal; NS, not significant; WT, wild type.
Comment in
- RNA sensing: the more RIG-I the merrier?
Rehwinkel J. Rehwinkel J. EMBO Rep. 2013 Sep;14(9):751-2. doi: 10.1038/embor.2013.120. Epub 2013 Aug 6. EMBO Rep. 2013. PMID: 23917614 Free PMC article. No abstract available.
Similar articles
- RIG-I ATPase activity and discrimination of self-RNA versus non-self-RNA.
Anchisi S, Guerra J, Garcin D. Anchisi S, et al. mBio. 2015 Mar 3;6(2):e02349. doi: 10.1128/mBio.02349-14. mBio. 2015. PMID: 25736886 Free PMC article. - Differential recognition of viral RNA by RIG-I.
Baum A, García-Sastre A. Baum A, et al. Virulence. 2011 Mar-Apr;2(2):166-9. doi: 10.4161/viru.2.2.15481. Epub 2011 Mar 1. Virulence. 2011. PMID: 21422808 Free PMC article. - ATP hydrolysis by the viral RNA sensor RIG-I prevents unintentional recognition of self-RNA.
Lässig C, Matheisl S, Sparrer KM, de Oliveira Mann CC, Moldt M, Patel JR, Goldeck M, Hartmann G, García-Sastre A, Hornung V, Conzelmann KK, Beckmann R, Hopfner KP. Lässig C, et al. Elife. 2015 Nov 26;4:e10859. doi: 10.7554/eLife.10859. Elife. 2015. PMID: 26609812 Free PMC article. - Links between recognition and degradation of cytoplasmic viral RNA in innate immune response.
Oshiumi H, Mifsud EJ, Daito T. Oshiumi H, et al. Rev Med Virol. 2016 Mar;26(2):90-101. doi: 10.1002/rmv.1865. Epub 2015 Dec 8. Rev Med Virol. 2016. PMID: 26643446 Review. - [Innate immune responses against viral infection and its suppression by viral proteins].
Oshiumi H, Matsumoto M, Seya T. Oshiumi H, et al. Yakugaku Zasshi. 2013;133(3):323-8. doi: 10.1248/yakushi.12-00237-5. Yakugaku Zasshi. 2013. PMID: 23449408 Review. Japanese.
Cited by
- Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor.
Zhu J, Zhang Y, Ghosh A, Cuevas RA, Forero A, Dhar J, Ibsen MS, Schmid-Burgk JL, Schmidt T, Ganapathiraju MK, Fujita T, Hartmann R, Barik S, Hornung V, Coyne CB, Sarkar SN. Zhu J, et al. Immunity. 2014 Jun 19;40(6):936-48. doi: 10.1016/j.immuni.2014.05.007. Epub 2014 Jun 12. Immunity. 2014. PMID: 24931123 Free PMC article. - The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly.
Bruns AM, Leser GP, Lamb RA, Horvath CM. Bruns AM, et al. Mol Cell. 2014 Sep 4;55(5):771-81. doi: 10.1016/j.molcel.2014.07.003. Epub 2014 Aug 7. Mol Cell. 2014. PMID: 25127512 Free PMC article. - The influenza virus RNA polymerase as an innate immune agonist and antagonist.
Elshina E, Te Velthuis AJW. Elshina E, et al. Cell Mol Life Sci. 2021 Dec;78(23):7237-7256. doi: 10.1007/s00018-021-03957-w. Epub 2021 Oct 22. Cell Mol Life Sci. 2021. PMID: 34677644 Free PMC article. Review. - What Really Rigs Up RIG-I?
Barik S. Barik S. J Innate Immun. 2016;8(5):429-36. doi: 10.1159/000447947. Epub 2016 Jul 21. J Innate Immun. 2016. PMID: 27438016 Free PMC article. Review. - Regulation of cGAS- and RLR-mediated immunity to nucleic acids.
Ablasser A, Hur S. Ablasser A, et al. Nat Immunol. 2020 Jan;21(1):17-29. doi: 10.1038/s41590-019-0556-1. Epub 2019 Dec 9. Nat Immunol. 2020. PMID: 31819255 Review.
References
- Ranjan P, Bowzard JB, Schwerzmann JW, Jeisy-Scott V, Fujita T, Sambhara S (2009) Cytoplasmic nucleic acid sensors in antiviral immunity. Trends Mol Med 15: 359–368 - PubMed
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T (2004) The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5: 730–737 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources