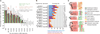A compendium of RNA-binding motifs for decoding gene regulation - PubMed (original) (raw)
. 2013 Jul 11;499(7457):172-7.
doi: 10.1038/nature12311.
Hilal Kazan, Kate B Cook, Matthew T Weirauch, Hamed S Najafabadi, Xiao Li, Serge Gueroussov, Mihai Albu, Hong Zheng, Ally Yang, Hong Na, Manuel Irimia, Leah H Matzat, Ryan K Dale, Sarah A Smith, Christopher A Yarosh, Seth M Kelly, Behnam Nabet, Desirea Mecenas, Weimin Li, Rakesh S Laishram, Mei Qiao, Howard D Lipshitz, Fabio Piano, Anita H Corbett, Russ P Carstens, Brendan J Frey, Richard A Anderson, Kristen W Lynch, Luiz O F Penalva, Elissa P Lei, Andrew G Fraser, Benjamin J Blencowe, Quaid D Morris, Timothy R Hughes
Affiliations
- PMID: 23846655
- PMCID: PMC3929597
- DOI: 10.1038/nature12311
A compendium of RNA-binding motifs for decoding gene regulation
Debashish Ray et al. Nature. 2013.
Abstract
RNA-binding proteins are key regulators of gene expression, yet only a small fraction have been functionally characterized. Here we report a systematic analysis of the RNA motifs recognized by RNA-binding proteins, encompassing 205 distinct genes from 24 diverse eukaryotes. The sequence specificities of RNA-binding proteins display deep evolutionary conservation, and the recognition preferences for a large fraction of metazoan RNA-binding proteins can thus be inferred from their RNA-binding domain sequence. The motifs that we identify in vitro correlate well with in vivo RNA-binding data. Moreover, we can associate them with distinct functional roles in diverse types of post-transcriptional regulation, enabling new insights into the functions of RNA-binding proteins both in normal physiology and in human disease. These data provide an unprecedented overview of RNA-binding proteins and their targets, and constitute an invaluable resource for determining post-transcriptional regulatory mechanisms in eukaryotes.
Figures
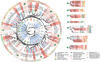
Figure 1. RNAcompete data for 207 RBPs
a, 7-mer Z scores and motifs for the two probe sets for ZC3H10. b, Two-dimensional hierarchical clustering analysis (Pearson correlation, average linkage) of E scores for 7-mers with E ≥ 0.4 in at least one experiment, with the two halves of the array kept as separate rows. Long systematic names have been shortened to species abbreviations and RNAcompete assay numbers. c, ROC curves showing discrimination of bound and unbound RNAs by the corresponding protein in vivo. The curve with the highest AUROC is shown if there are multiple in vivo data sets for a protein. FUS and TAF15 were excluded.
Figure 2. Motifs obtained by RNAcompete for RRM (outer ring) and KH domain proteins (inner ring)
The dendrograms represent complete linkage hierarchical clustering of RBPs by amino acid sequence identity in their RBDs. Line colours indicate species of origin of each protein, and shading indicates clades in which all sequences are more than 70% (dark) or 50% (light) identical.
Figure 3. RBD sequence identity enables inference of RNA motifs
a, Motif similarity versus per cent amino acid sequence identity in all RBDs for pairs of proteins. Motif similarity scored using STAMP Pearson-based log10(E value), correlation between PFM affinity scores against 10,000 random-sequence 100-mers, or human 3′ UTRs (for human RBPs). Columns indicate average; error bars indicate standard deviation. Red points: new proteins analysed (see c). b, Stacked bars indicate proportion of each category of RBP encompassed by experimentally determined motifs or inferred motifs using stringent (RNAcompete motifs, ≥70% identity) or expanded criteria (RNAcompete and literature motifs, ≥50% identity) in 288 eukaryotes (Supplementary Data 9). ‘Multi-RBD’ and “All” indicate proteins with >1 or >0 RBDs, respectively. c, Validation of motifs predicted for proteins at 61–96% amino acid identity (red text indicates validation motifs).
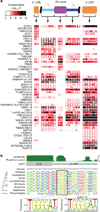
Figure 4. Conservation of motif matches in human RNA regulatory regions
a, Heat map showing conservation in 50-nucleotide bins (columns) in regions indicated at the top of the panel. Rows represent the most significant motif for indicated protein family (see Supplementary Table 4). Box fill: conservation score of the most conserved position in the motif for each bin. Border colour: conservation score when the entire regulatory region is considered as a single bin. Asterisks indicate known splicing factors. b, Alignment of vertebrate sequences over the ESRP1/2 site in the USF1 3′ UTR. Sequence logos are shown for major branches of vertebrate taxonomy. Dashed box: motif derived from the full alignment. The RNAcompete motif for ESRP1/2 is shown to the right.
Figure 5. RBFOX1 is a putative regulator of RNA stability in autism
a, Significance (as rank-sum Z score) of bias that RBP motifs in 3′ UTRs of mRNAs confer towards correlated expression with the RBP’s mRNA (FDR <0.1). b, Scatter plot shows Z score (from a) versus rank-sum Z score of the same target set, with mRNAs ranked instead by decay rate in MDA-MB-231 cells, for expressed RBPs. c, Enrichment of predicted RBFOX1 stability targets (by ‘leading-edge’ analysis) among transcripts with conserved RBFOX1 motifs. d, Density plot showing that RBFOX1 targets are enriched among transcripts most affected by RBFOX1 RNAi. e, Relationship of mRNA expression levels in autism spectrum disorder brains to RBFOX1 expression and predicted RBFOX1 target status.
Similar articles
- Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs.
Melé M, Mattioli K, Mallard W, Shechner DM, Gerhardinger C, Rinn JL. Melé M, et al. Genome Res. 2017 Jan;27(1):27-37. doi: 10.1101/gr.214205.116. Epub 2016 Dec 7. Genome Res. 2017. PMID: 27927715 Free PMC article. - ELAV multimerizes on conserved AU4-6 motifs important for ewg splicing regulation.
Soller M, White K. Soller M, et al. Mol Cell Biol. 2005 Sep;25(17):7580-91. doi: 10.1128/MCB.25.17.7580-7591.2005. Mol Cell Biol. 2005. PMID: 16107705 Free PMC article. - A deep boosting based approach for capturing the sequence binding preferences of RNA-binding proteins from high-throughput CLIP-seq data.
Li S, Dong F, Wu Y, Zhang S, Zhang C, Liu X, Jiang T, Zeng J. Li S, et al. Nucleic Acids Res. 2017 Aug 21;45(14):e129. doi: 10.1093/nar/gkx492. Nucleic Acids Res. 2017. PMID: 28575488 Free PMC article. - Protein Binding to Cis-Motifs in mRNAs Coding Sequence Is Common and Regulates Transcript Stability and the Rate of Translation.
Grzybowska EA, Wakula M. Grzybowska EA, et al. Cells. 2021 Oct 27;10(11):2910. doi: 10.3390/cells10112910. Cells. 2021. PMID: 34831133 Free PMC article. Review. - lncRNAs in development and differentiation: from sequence motifs to functional characterization.
Constanty F, Shkumatava A. Constanty F, et al. Development. 2021 Jan 13;148(1):dev182741. doi: 10.1242/dev.182741. Development. 2021. PMID: 33441380 Review.
Cited by
- A bioinformatic approach for the prediction and functional classification of Toxoplasma gondii long non-coding RNAs.
Vanagas L, Cristaldi C, La Bella G, Ganuza A, Angel SO, Alonso AM. Vanagas L, et al. Sci Rep. 2024 Nov 12;14(1):27687. doi: 10.1038/s41598-024-79204-6. Sci Rep. 2024. PMID: 39533086 Free PMC article. - Circular RNA landscape in extracellular vesicles from human biofluids.
Zhao J, Li Q, Hu J, Yu H, Shen Y, Lai H, Li Q, Zhang H, Li Y, Fang Z, Huang S. Zhao J, et al. Genome Med. 2024 Oct 31;16(1):126. doi: 10.1186/s13073-024-01400-w. Genome Med. 2024. PMID: 39482783 Free PMC article. - The MTR4/hnRNPK complex surveils aberrant polyadenylated RNAs with multiple exons.
Taniue K, Sugawara A, Zeng C, Han H, Gao X, Shimoura Y, Ozeki AN, Onoguchi-Mizutani R, Seki M, Suzuki Y, Hamada M, Akimitsu N. Taniue K, et al. Nat Commun. 2024 Oct 17;15(1):8684. doi: 10.1038/s41467-024-51981-8. Nat Commun. 2024. PMID: 39419981 Free PMC article. - RNAelem: an algorithm for discovering sequence-structure motifs in RNA bound by RNA-binding proteins.
Miyake H, Kawaguchi RK, Kiryu H. Miyake H, et al. Bioinform Adv. 2024 Sep 28;4(1):vbae144. doi: 10.1093/bioadv/vbae144. eCollection 2024. Bioinform Adv. 2024. PMID: 39399375 Free PMC article. - PTBP1 mediates Sertoli cell actin cytoskeleton organization by regulating alternative splicing of actin regulators.
Wang Y, Chembazhi UV, Yee D, Chen S, Ji J, Wang Y, Nguyen KL, Lin P, Ratti A, Hess RA, Qiao H, Ko C, Yang J, Kalsotra A, Mei W. Wang Y, et al. Nucleic Acids Res. 2024 Nov 11;52(20):12244-12261. doi: 10.1093/nar/gkae862. Nucleic Acids Res. 2024. PMID: 39373517 Free PMC article.
References
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nature Rev. Genet. 2007;8:533–543. - PubMed
- Gabut M, Chaudhry S, Blencowe BJ. SnapShot: The splicing regulatory machinery. Cell. 2008;133:192.e1. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK015602-05/DK/NIDDK NIH HHS/United States
- MOP-93671/CAPMC/ CIHR/Canada
- R01 CA104708/CA/NCI NIH HHS/United States
- MOP-49451/CAPMC/ CIHR/Canada
- P30 CA014520/CA/NCI NIH HHS/United States
- R01 GM051968/GM/NIGMS NIH HHS/United States
- MOP-14409/CAPMC/ CIHR/Canada
- 1R01HG00570/HG/NHGRI NIH HHS/United States
- T32 GM008061/GM/NIGMS NIH HHS/United States
- R01 GM084034/GM/NIGMS NIH HHS/United States
- R01 GM058728/GM/NIGMS NIH HHS/United States
- MOP-125894/CAPMC/ CIHR/Canada
- R01GM084034/GM/NIGMS NIH HHS/United States
- MOP-67011/CAPMC/ CIHR/Canada
- Z01 DK015602-01/ImNIH/Intramural NIH HHS/United States
- R01 GM114386/GM/NIGMS NIH HHS/United States
- R01 HG005700/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases