TRiC's tricks inhibit huntingtin aggregation - PubMed (original) (raw)
TRiC's tricks inhibit huntingtin aggregation
Sarah H Shahmoradian et al. Elife. 2013.
Abstract
In Huntington's disease, a mutated version of the huntingtin protein leads to cell death. Mutant huntingtin is known to aggregate, a process that can be inhibited by the eukaryotic chaperonin TRiC (TCP1-ring complex) in vitro and in vivo. A structural understanding of the genesis of aggregates and their modulation by cellular chaperones could facilitate the development of therapies but has been hindered by the heterogeneity of amyloid aggregates. Using cryo-electron microscopy (cryoEM) and single particle cryo-electron tomography (SPT) we characterize the growth of fibrillar aggregates of mutant huntingtin exon 1 containing an expanded polyglutamine tract with 51 residues (mhttQ51), and resolve 3-D structures of the chaperonin TRiC interacting with mhttQ51. We find that TRiC caps mhttQ51 fibril tips via the apical domains of its subunits, and also encapsulates smaller mhtt oligomers within its chamber. These two complementary mechanisms provide a structural description for TRiC's inhibition of mhttQ51 aggregation in vitro. DOI:http://dx.doi.org/10.7554/eLife.00710.001\.
Keywords: Amyloid; Cryo electron microscopy (cryoEM); Cryo electron tomography (cryoET); Huntingtin; None; Single particle tomography (SPT); TRiC chaperonin.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
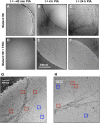
Figure 1.. The presence of TRiC inhibits the progression of mhttQ51 aggregation.
2-D cryoEM images of (A) mhttQ51 at 45 min, (B) 4 hr, and (C) 24 hr post-initiation of aggregation (PIA), and of mhttQ51 incubated with TRiC and imaged at (D) 45 min (E) 4 hr and (F) 24 hr PIA. (G, H) Higher magnification images of mhttQ51 + TRiC at 4 hr PIA. Some TRiCs are seemingly attached to mhtt fibrils (red boxes), while others are seemingly freestanding (blue boxes). DOI:
http://dx.doi.org/10.7554/eLife.00710.003
Figure 2.. TRiC caps the tips of mhttQ51 fibrils.
(A) 2-D slice through a tomogram of TRiC incubated with mhttQ51 imaged at 4 hr post initiation of aggregation. (B) 2-D reprojection of the entire tomogram and (C) 3-D annotation of its features, showing mhtt fibrils in yellow, freestanding TRiCs in blue, and fibril-bound TRiCs in magenta. Zoomed-in field shows fibril-bound TRiC localizing mostly to discernible tips of mhtt fibrils (seemingly detached magenta TRiC is attached to a fibril on another z-slice of the tomogram). DOI:
http://dx.doi.org/10.7554/eLife.00710.004
Figure 3.. TRiC’s apical tips bind mhtt-fibrils.
SPT symmetry-free and model-free averages of fibril-bound TRiC reveal extra density at the chaperonin’s apical tips. Each panel shows an end-on and a side view isosurface for a 3-D average of TRiC in complex with mhttQ51 fibrils, and corresponding 2-D projections (black/white). Independent sets from single tomograms are labeled A, B, C, D and E. Within each, the numbering (i, ii and iii) indicates mutually exclusive averages. The numbers of subvolumes contributing to each average are (A.i.) 7, (A.ii.) 10, (A.iii.) 13, (B.i.) 7, (B.ii.) 10, (B.iii.) 5, (C) 12, (D) 8 and (E) 12. All the averages show overall TRiC-like morphology with extra densities protruding beyond the apical domains of the chaperonin. DOI:
http://dx.doi.org/10.7554/eLife.00710.006
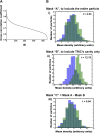
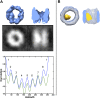
Figure 4.. Freestanding TRiCs are classifiable into cavity-empty and cavity-occupied.
(A) Ranked correlation scores for freestanding TRiCs against a hollow TRiC reference yield a continuous fast-decaying plot, suggesting structural heterogeneity. (B) Mean-density distributions for cavity-empty TRiC (blue) and cavity-occupied TRiC (green) sets, masking the particles to (i) include all the density in each particle, (ii) include the cavity of each molecule only, and (iii) include TRiC density but exclude the cavity. Corresponding _t_-scores for the difference (or shift) between the blue and green distributions are shown for each masking condition. DOI:
http://dx.doi.org/10.7554/eLife.00710.007
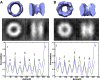
Figure 5.. A subpopulation of freestanding TRiC contains intra-cavity extra density.
SPT symmetry-free and template-free processing of freestanding TRiC demonstrates pseudo-eightfold symmetry in an average of (A) 356 cavity-empty TRiCs and (B) 356 cavity-occupied TRiCs. 2-D projections for each average are shown in black/white. The plots show volumetric rotational correlation analyses to test for C8 (blue) and D8 (green) symmetry in each average. The cavity size in B is significantly reduced, due to internalized density. DOI:
http://dx.doi.org/10.7554/eLife.00710.008
Figure 6.. Control TRiC in the absence of mhtt.
End-on and cut-away views for (A) the published cryoEM structure of apo-TRiC (EMDB 1960), low-pass filtered to match the resolution of freestanding TRiCs, and (B) model-free SPT TRiC average of 208 subvolumes from TRiC-only tomograms or ‘control TRiC’ (no mhtt added). Corresponding projections for the control are shown. DOI:
http://dx.doi.org/10.7554/eLife.00710.009
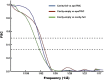
Figure 7.. FSC curves for freestanding TRiC averages.
FSC of cavity-empty TRiC (maroon) and cavity-occupied TRiC (blue) with the published structure of apo-TRiC. The FSC of cavity-empty TRiC with cavity-occupied TRiC is shown in green. DOI:
http://dx.doi.org/10.7554/eLife.00710.010
Figure 8.. Localized intra-cavity extra density within TRiC.
(A) Hollow-template-guided average of 356 cavity-occupied TRiCs, 2-D projections (black/white) and rotational correlation analysis to test for C8 (blue) and D8 (green) symmetry, showing a ‘dip’ in self-correlation at azimuth equal to 180° due to the presence of localized inner mass. (B) End-on (left) and cut-away side (right) views of the superimposition of the cavity-empty TRiC average (blue wire) from Figure 5A, and the difference map (yellow) between it and the cavity-occupied average in (A). DOI:
http://dx.doi.org/10.7554/eLife.00710.011
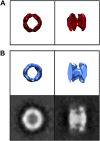
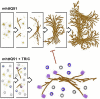
Figure 9.. Model for the progression of mhttQ51 aggregation in the absence and presence of TRiC.
In the absence of TRiC (top row), mhttQ51 (orange) aggregation progresses from monomers and oligomers into fibrils and large sheaves. On the other hand, the presence of TRiC (bottom row) prevents the progression of mhtt fibrils into the large bundled ‘sheaf’ form by capping (purple) fibril tips and encapsulating (blue) mhtt oligomers. TRiCs that are not bound to mhtt are shown in gray. DOI:
http://dx.doi.org/10.7554/eLife.00710.012
Similar articles
- Structural Mechanisms of Mutant Huntingtin Aggregation Suppression by the Synthetic Chaperonin-like CCT5 Complex Explained by Cryoelectron Tomography.
Darrow MC, Sergeeva OA, Isas JM, Galaz-Montoya JG, King JA, Langen R, Schmid MF, Chiu W. Darrow MC, et al. J Biol Chem. 2015 Jul 10;290(28):17451-61. doi: 10.1074/jbc.M115.655373. Epub 2015 May 20. J Biol Chem. 2015. PMID: 25995452 Free PMC article. - 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement.
Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, Reissmann S, Kumar RN, Redding-Johanson AM, Batth TS, Mukhopadhyay A, Ludtke SJ, Frydman J, Chiu W. Cong Y, et al. Proc Natl Acad Sci U S A. 2010 Mar 16;107(11):4967-72. doi: 10.1073/pnas.0913774107. Epub 2010 Mar 1. Proc Natl Acad Sci U S A. 2010. PMID: 20194787 Free PMC article. - Exogenous delivery of chaperonin subunit fragment ApiCCT1 modulates mutant Huntingtin cellular phenotypes.
Sontag EM, Joachimiak LA, Tan Z, Tomlinson A, Housman DE, Glabe CG, Potkin SG, Frydman J, Thompson LM. Sontag EM, et al. Proc Natl Acad Sci U S A. 2013 Feb 19;110(8):3077-82. doi: 10.1073/pnas.1222663110. Epub 2013 Jan 30. Proc Natl Acad Sci U S A. 2013. PMID: 23365139 Free PMC article. - Huntingtin-protein interactions and the pathogenesis of Huntington's disease.
Li SH, Li XJ. Li SH, et al. Trends Genet. 2004 Mar;20(3):146-54. doi: 10.1016/j.tig.2004.01.008. Trends Genet. 2004. PMID: 15036808 Review. - Are there multiple pathways in the pathogenesis of Huntington's disease?
Aronin N, Kim M, Laforet G, DiFiglia M. Aronin N, et al. Philos Trans R Soc Lond B Biol Sci. 1999 Jun 29;354(1386):995-1003. doi: 10.1098/rstb.1999.0451. Philos Trans R Soc Lond B Biol Sci. 1999. PMID: 10434298 Free PMC article. Review.
Cited by
- Contribution of Extracellular Vesicles and Molecular Chaperones in Age-Related Neurodegenerative Disorders of the CNS.
Noori L, Filip K, Nazmara Z, Mahakizadeh S, Hassanzadeh G, Caruso Bavisotto C, Bucchieri F, Marino Gammazza A, Cappello F, Wnuk M, Scalia F. Noori L, et al. Int J Mol Sci. 2023 Jan 4;24(2):927. doi: 10.3390/ijms24020927. Int J Mol Sci. 2023. PMID: 36674442 Free PMC article. Review. - Chaperonin-containing TCP-1 subunit 2-mediated aggrephagy: A potential target for treating neurodegeneration.
Chen X, Zhang M. Chen X, et al. Clin Transl Med. 2022 Aug;12(8):e1027. doi: 10.1002/ctm2.1027. Clin Transl Med. 2022. PMID: 35988155 Free PMC article. No abstract available. - Hidden motions and motion-induced invisibility: Dynamics-based spectral editing in solid-state NMR.
Matlahov I, van der Wel PCA. Matlahov I, et al. Methods. 2018 Sep 15;148:123-135. doi: 10.1016/j.ymeth.2018.04.015. Epub 2018 Apr 24. Methods. 2018. PMID: 29702226 Free PMC article. Review. - Missing Wedge Completion via Unsupervised Learning with Coordinate Networks.
Van Veen D, Galaz-Montoya JG, Shen L, Baldwin P, Chaudhari AS, Lyumkis D, Schmid MF, Chiu W, Pauly J. Van Veen D, et al. Int J Mol Sci. 2024 May 17;25(10):5473. doi: 10.3390/ijms25105473. Int J Mol Sci. 2024. PMID: 38791508 Free PMC article. - Protofilament Structure and Supramolecular Polymorphism of Aggregated Mutant Huntingtin Exon 1.
Boatz JC, Piretra T, Lasorsa A, Matlahov I, Conway JF, van der Wel PCA. Boatz JC, et al. J Mol Biol. 2020 Jul 24;432(16):4722-4744. doi: 10.1016/j.jmb.2020.06.021. Epub 2020 Jun 27. J Mol Biol. 2020. PMID: 32598938 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- PN2EY016525/EY/NEI NIH HHS/United States
- T32 EB009379/EB/NIBIB NIH HHS/United States
- PN2 EY016525/EY/NEI NIH HHS/United States
- R01 GM080139/GM/NIGMS NIH HHS/United States
- R01GM080139/GM/NIGMS NIH HHS/United States
- P41GM103832/GM/NIGMS NIH HHS/United States
- P41 GM103832/GM/NIGMS NIH HHS/United States
- T32EB009379/EB/NIBIB NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous