BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival - PubMed (original) (raw)
. 2013 Jul 29;210(8):1529-44.
doi: 10.1084/jem.20121337. Epub 2013 Jul 15.
Pegah S Baniasadi, Isaac S Harris, Jennifer Silvester, Satoshi Inoue, Bryan Snow, Purna A Joshi, Andrew Wakeham, Sam D Molyneux, Bernard Martin, Peter Bouwman, David W Cescon, Andrew J Elia, Zoe Winterton-Perks, Jennifer Cruickshank, Dirk Brenner, Alan Tseng, Melinda Musgrave, Hal K Berman, Rama Khokha, Jos Jonkers, Tak W Mak, Mona L Gauthier
Affiliations
- PMID: 23857982
- PMCID: PMC3727320
- DOI: 10.1084/jem.20121337
BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival
Chiara Gorrini et al. J Exp Med. 2013.
Abstract
Oxidative stress plays an important role in cancer development and treatment. Recent data implicate the tumor suppressor BRCA1 in regulating oxidative stress, but the molecular mechanism and the impact in BRCA1-associated tumorigenesis remain unclear. Here, we show that BRCA1 regulates Nrf2-dependent antioxidant signaling by physically interacting with Nrf2 and promoting its stability and activation. BRCA1-deficient mouse primary mammary epithelial cells show low expression of Nrf2-regulated antioxidant enzymes and accumulate reactive oxygen species (ROS) that impair survival in vivo. Increased Nrf2 activation rescues survival and ROS levels in BRCA1-null cells. Interestingly, 53BP1 inactivation, which has been shown to alleviate several defects associated with BRCA1 loss, rescues survival of BRCA1-null cells without restoring ROS levels. We demonstrate that estrogen treatment partially restores Nrf2 levels in the absence of BRCA1. Our data suggest that Nrf2-regulated antioxidant response plays a crucial role in controlling survival downstream of BRCA1 loss. The ability of estrogen to induce Nrf2 posits an involvement of an estrogen-Nrf2 connection in BRCA1 tumor suppression. Lastly, BRCA1-mutated tumors retain a defective antioxidant response that increases the sensitivity to oxidative stress. In conclusion, the role of BRCA1 in regulating Nrf2 activity suggests important implications for both the etiology and treatment of BRCA1-related cancers.
Figures
Figure 1.
BRCA1 loss-of-function induces high ROS levels in MECs. (A) Brca1 mRNA expression in COMMA-1D cells infected with dox-inducible Brca1 shRNA and treated (+dox) or not (−dox) with dox. (B) Quantitation of ROS in COMMA-1D cells as processed in A. (C) BRCA1 mRNA levels in HMECs infected with Luc shRNA (control) or BRCA1 shRNA. (D) Quantitation of ROS in HMEC as treated in C. (A–D) Data represent the mean ± SEM of three biological replicates. (E) Representative PCR with genomic DNA isolated from B1+/+, B1f/+, B1f/f, and KB1f/f pMECs using specific primers for detection of Brca1 WT allele, loxP site in intron 3 (F), or cre-mediated deleted allele (Δ). Primers are described in Liu et al. (2007) and
Table S1
. (F) qPCR with genomic DNA from K, KB1f/+, and KB1f/f pMECs using specific primers directed against Brca1 WT allele as reported in Table S1. (G) BRCA1 mRNA levels in K, KB1f/+, and KB1f/f pMECs. (H) Representative analysis of BRCA1 protein levels in K and KB1f/f pMECs. Vinculin was used as a loading control. (I) ROS levels in K, KB1f/+, and KB1f/f pMECs. (F, G, and I) Data represent the mean ± SEM of n = 5 mice of each genotype. (J) Representative FACS profile of ROS levels in MaSC/basal and luminal cell subpopulations in B1f/f and KB1f/f pMECs stained with DCF-DA. *, P < 0.05; **, P < 0.01.
Figure 2.
Brca1 expression is regulated by oxidative stress and controls Nrf2 abundance and transactivation activity. (A) Nrf2 mRNA levels in B1f/f and KB1f/f pMECs. (B) Representative immunoblot of Nrf2 protein levels in pMECs from B1f/f (n = 2) and KB1f/f (n = 3) mice. Western blot quantitation is shown below. (C and D) RT-PCR analysis of Nrf2 targets, Nqo1 (C) and Hmox1 (D), in B1f/f and KB1f/f pMECs. (A, C, and D) Data are the mean ± SEM of n = 5 mice of each genotype. (E) Representative immunoblot of Nrf2 protein levels in control (Con) or BSO-treated COMMA-1D cells after transfection with scrambled siRNA (siScr) or Brca1-specific siRNA (siBrca1). (F) Nrf2 mRNA levels in cells treated as in E. (G) Expression levels of Nqo1, Hmox1, and Brca1 mRNAs in cells treated as in E. (H) Representative immunoblot of Brca1 protein levels in control and BSO-treated COMMA-1D cells. Vinculin was used as a loading control. (I) Representative cell cycle profile by PI staining of control or BSO (1 or 2 mM)-treated COMMA-1D cells. (J) Brca1 mRNA levels in COMMA-1D cells that have been left untreated (Con) or after treatment with Trolox, BSO, and Trolox plus BSO for 48 h. (K) Nqo1 and Brca1 mRNA expression in untreated COMMA-1D cells transfected with scrambled siRNA or Nrf2-specific siRNA (siNrf2). (F, G, J, and K) Data represent the mean ± SEM of three biological replicates. *, P < 0.05; **, P < 0.01.
Figure 3.
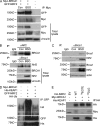
BRCA1 physically interacts with Nrf2 and affects Keap1-mediated Nrf2 ubiquitination. (A) 293FT cells were transfected with Myc-BRCA1 and GFP-NRF2 constructs and then left untreated (Con) or treated with BSO. BRCA1 was immunoprecipitated with anti-Myc Ab. The blot was probed with anti-Myc and anti-GFP Abs. “−” indicates untreated EV control. Vinculin was used as a loading control. (B) COMMA-1D cells were left untreated (Con) or treated with BSO and processed for immunoprecipitation (IP) with anti-Nrf2 (H-300) or anti-Brca1 (C20) Abs to detect the endogenous Brca1 and Nrf2 proteins. (C) COMMA-1D cells were treated as in B and subjected to immunoprecipitation with anti–mouse Brca1 Ab to detect endogenous Brca1 and Nrf2 complex. Nrf2 was detected with an affinity-purified Ab as described in Materials and methods. (B and C) IgG served as an isotype control. (D) 293FT cells were transfected with constructs expressing HA-ubiquitin, His-KEAP1, GFP-NRF2, and/or Myc-BRCA1 (as indicated). Immunoprecipitation was performed with anti-GFP Ab followed by Western blot to detect ubiquitinated NRF2 (HA, GFP) and His-KEAP1. (E) 293FT cells were transfected with Myc-BRCA1, His-KEAP1, HA–WT NRF2 (WT), HA–79EGTE NRF2 mutant (79EGTE), and HA–29DLG NRF2 mutant (29DLG). Immunoprecipitation was performed with anti-HA Ab, and Western blot was probed with anti-Myc and anti-His Abs. The asterisk indicates an unspecific band.
Figure 4.
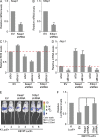
Brca1-deficient pMECs can regenerate a functional mammary gland but have a limited lifespan in vivo. (A) Breeding strategy used to obtain B1f/fLucf/+, KLucf/+, and KB1f/fLucf/+ animals used for in vivo fat pad transplantation assays. pMECs isolated from B1f/fLucf/+, KLucf/+, and KB1f/fLucf/+ donor mice were transplanted in 21-wk-old recipient mice to generate outgrowths. (B) In vivo Luc activity in outgrowths derived from B1f/fLucf/+ (mouse 1 as negative control), KLucf/+ (mice 2 and 8 as positive controls), and KB1f/fLucf/+ (mice 3–7) pMECs at 4 and 24 wk after transplantation. (C) Quantitation of Luc activity shown in B using Living Image 3.0 software. (D) qPCR of Brca1 WT allele with genomic DNA from pMECs isolated from KLucf/+ and KB1f/fLucf/+ 4-wk outgrowths. (E) Immunohistochemical staining of 4-wk KLucf/+ and KB1f/fLucf/+ outgrowths with anti-Luc and anti-CK14 Abs. Phase-contrast images of the same glands are shown below. (F) Whole-mount staining of KLucf/+ and KB1f/fLucf/+ 4-wk outgrowths. Terminal end buds are shown at higher magnification. (G) Hematoxylin-eosin staining of KLucf/+ and KB1f/fLucf/+ outgrowths at 24 wk after transplantation. Data are representative of 10 outgrowths examined per genotype. Bars: (E) 16 µm; (F and G) 50 µm. (H and I) RT-PCR analysis of p16/INK4 (H) and p19/ARF (I) mRNA levels in KLucf/+ and KB1f/fLucf/+ 8-wk outgrowths. (J) pMECs from KLucf/+ and KB1f/fLucf/+ 8-wk outgrowths were stained with FITC-γH2AX and analyzed by flow cytometry. (K) p21/Cdkn1a mRNA levels in pMECs from KLucf/+ and KB1f/fLucf/+ 8-wk outgrowths. (C, D, and H–K) Data represent the mean ± SEM of n = 5 outgrowths of each genotype. *, P < 0.05; **, P < 0.01.
Figure 5.
Keap1 down-regulation rescues in vivo survival defect and ROS levels in BRCA1-deficient cells. (A) Keap1 mRNA levels in COMMA-1D cells infected with EV or Keap1 shRNA. (B) 53bp1 mRNA levels in COMMA-1D cells infected with EV or 53bp1 shRNA. (C) ROS levels in COMMA-1D cells stably expressing EV or Keap1 and 53bp1 shRNAs after transfection with scrambled siRNA (siScr) or Brca1-specific siRNA (siBrca1). Data are normalized to ROS levels in EV/siScr cells. (D) Nqo1 mRNA levels in cells as described in C. Data are normalized to siScr/EV cells. (A–D) Data represent the mean ± SEM of three biological replicates. (E) Representative images of in vivo Luc activity in outgrowths derived from KLucf/+ pMECs (mouse 1, positive control) and KB1f/fLucf/+ pMECs infected with EV (mouse 2), Keap1 shRNA (mice 3 and 4), or 53bp1 shRNA (mice 5 and 6). (F) Quantitation of Luc activity shown in E using Living Image 3.0 software. Data represent the mean ± SEM from n = 5 outgrowths of each combination. *, P < 0.05; **, P < 0.01.
Figure 6.
Estrogen stimulates an Nrf2-regulated antioxidant response. (A) Representative immunoblot analysis of NRF2 protein in estrogen (E2)-starved MCF7 cells that were treated with 10 nM E2 for 2 or 4 h. (B and C) MCF7 cells that were left untreated (Con) or treated as in A for 4 h were used to detect NRF2 (B) and GCLM (C) mRNA levels by RT-PCR. (D) Representative ROS analysis by FACS in MCF7 cells treated as in B. (E) Representative cell cycle analysis by PI staining of control (Con) or E2-treated MCF7 cells (10 nM for 4 h). (F) HC11 cells were left untreated or stimulated with different doses of E2 as indicated for 4-h Nrf2, and vinculin protein levels were detected. Representative Western blot is shown. (G) Gclm mRNA levels in HC11 treated with 10 nM E2 for 4 h. (B, C, and G) Data represent the mean ± SEM of three biological replicates. (H and I) RT-PCR analysis of Gclm (H) and Nqo1 (I) mRNA levels in luminal (L) and MaSC/basal (B) cells isolated from ovariectomized FVB mice that were treated with vehicle or 0.14 mg E2 for 14 d. Data represent the mean ± SEM of n = 3 independent isolates. L, B = luminal or MaSC/basal cells from vehicle-treated mice; L+E2, B+E2 = luminal or basal cells from E2-treated mice (n = 3/group). (J) Representative Western blot analysis of Nrf2 protein levels in B1f/f and KB1f/f pMECs treated with 10 nM E2 for 2 and 4 h. Vinculin was used as a loading control. *, P < 0.05; **, P < 0.01.
Figure 7.
Reconstitution of WT BRCA1 in a human _BRCA1_-mutated breast cancer cell line restores Nrf2 activation. (A) RT-PCR analysis of BRCA1 mRNA levels in HCCWT and HCCmut cells. (B) Quantitation of ROS levels analyzed by FACS in HCCWT and HCCmut cells. (C) Cell viability of HCCWT and HCCmut cells at 4 d after treatment with 5 mM BSO. Data are the mean percentage ± SEM of viable cells compared with untreated controls (n = 3/group). (D) Representative immunoblot of NRF2 protein levels in HCCWT and HCCmut cells that were left untreated (Con) or treated with the indicated doses of BSO. H3 served as a loading control. (E and F) NQO1 (E) and GCLM (F) expression levels in HCCWT and HCCmut cells transfected with scrambled siRNA (siScr) and KEAP1 siRNA (siKEAP1). (G) ROS levels in siScr- and siKeap1-transfected HCCWT and HCCmut cells. Data in siKeap1 cells are normalized to siScr controls for each cell line (n = 3/group). Data represent the mean ± SEM of three biological replicates, normalized to siScr-transfected cells. (H) Analysis of apoptosis by Annexin V/7-AAD staining of HCCWT and HCCmut cells left untreated (Con) or treated with 10 or 30 µM cisplatin for 24 h. (I) Analysis of apoptosis by Annexin V/7-AAD staining in siScr- and siKeap1-transfected HCCWT and HCCmut cells left untreated (Con) or treated with 10 µM cisplatin (CIS) for 24 h. Data are normalized to untreated siScr cells for each cell line. (A, B, E, F, H, and I) Data represent the mean ± SEM of three biological replicates. *, P < 0.05; **, P < 0.01.
Figure 8.
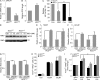
NQO1 is down-regulated in _Brca1_-null mouse mammary tumors and _BRCA1_-mutated human breast cancers. (A) Immunohistochemical analysis of Nqo1 protein in normal mouse mammary gland (left) and in a mammary tumor arising in a K14cre Brca1f/fp53f/f mouse (KB1f/fp53f/f; right). Results are representative of five glands from normal FVB mice and five tumors from KB1f/fp53f/f mice. Bars, 50 µm. (B–D) Box plots representing NQO1 mRNA levels in human primary _BRCA1_-mutated breast cancers (BRCA1mut) and _BRCA1_-proficient breast cancers (BRCA1WT) from three different datasets as indicated in each panel and described in Materials and methods. (E–G) Dot plots showing NQO1 mRNA levels in the indicated human primary breast tumor subtypes from three different datasets as indicated in each panel and described in Materials and methods. Vertical lines indicate the variability outside the upper and lower quartiles represented in the box plots. P-values were determined by Kruskal–Wallis analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.0001.
Comment in
- Refining the role of BRCA1 in combating oxidative stress.
Marks JR. Marks JR. Breast Cancer Res. 2013 Dec 5;15(6):320. doi: 10.1186/bcr3583. Breast Cancer Res. 2013. PMID: 24314328 Free PMC article.
Similar articles
- Continuous activation of Nrf2 and its target antioxidant enzymes leads to arsenite-induced malignant transformation of human bronchial epithelial cells.
Yang X, Wang D, Ma Y, Xu X, Zhu Z, Wang X, Deng H, Li C, Chen M, Tong J, Yamanaka K, An Y. Yang X, et al. Toxicol Appl Pharmacol. 2015 Dec 1;289(2):231-9. doi: 10.1016/j.taap.2015.09.020. Epub 2015 Sep 28. Toxicol Appl Pharmacol. 2015. PMID: 26420645 - Bioactive food components prevent carcinogenic stress via Nrf2 activation in BRCA1 deficient breast epithelial cells.
Kang HJ, Hong YB, Kim HJ, Wang A, Bae I. Kang HJ, et al. Toxicol Lett. 2012 Mar 7;209(2):154-60. doi: 10.1016/j.toxlet.2011.12.002. Epub 2011 Dec 13. Toxicol Lett. 2012. PMID: 22192953 Free PMC article. - Estrogen controls the survival of BRCA1-deficient cells via a PI3K-NRF2-regulated pathway.
Gorrini C, Gang BP, Bassi C, Wakeham A, Baniasadi SP, Hao Z, Li WY, Cescon DW, Li YT, Molyneux S, Penrod N, Lupien M, Schmidt EE, Stambolic V, Gauthier ML, Mak TW. Gorrini C, et al. Proc Natl Acad Sci U S A. 2014 Mar 25;111(12):4472-7. doi: 10.1073/pnas.1324136111. Epub 2014 Feb 24. Proc Natl Acad Sci U S A. 2014. PMID: 24567396 Free PMC article. - BRCA1 and estrogen/estrogen receptor in breast cancer: where they interact?
Wang L, Di LJ. Wang L, et al. Int J Biol Sci. 2014 May 14;10(5):566-75. doi: 10.7150/ijbs.8579. eCollection 2014. Int J Biol Sci. 2014. PMID: 24910535 Free PMC article. Review. - Simultaneous Activation of Nrf2 and Elevation of Dietary and Endogenous Antioxidant Chemicals for Cancer Prevention in Humans.
Prasad KN. Prasad KN. J Am Coll Nutr. 2016;35(2):175-84. doi: 10.1080/07315724.2014.1003419. Epub 2015 Jul 7. J Am Coll Nutr. 2016. PMID: 26151600 Review.
Cited by
- The interplay between microRNAs and Nrf2 signaling in human cancers.
Panahizadeh R, Vatankhah MA, Safari A, Danesh H, Nazmi N, Gholizadeh P, Soozangar N, Jeddi F. Panahizadeh R, et al. Cancer Cell Int. 2024 Jul 5;24(1):234. doi: 10.1186/s12935-024-03430-1. Cancer Cell Int. 2024. PMID: 38970040 Free PMC article. Review. - Reactive Oxygen Species: the Dual Role in Physiological and Pathological Conditions of the Human Body.
Bardaweel SK, Gul M, Alzweiri M, Ishaqat A, ALSalamat HA, Bashatwah RM. Bardaweel SK, et al. Eurasian J Med. 2018 Oct;50(3):193-201. doi: 10.5152/eurasianjmed.2018.17397. Eurasian J Med. 2018. PMID: 30515042 Free PMC article. Review. - Interaction of ERα and NRF2 Impacts Survival in Ovarian Cancer Patients.
Czogalla B, Kahaly M, Mayr D, Schmoeckel E, Niesler B, Kolben T, Burges A, Mahner S, Jeschke U, Trillsch F. Czogalla B, et al. Int J Mol Sci. 2018 Dec 29;20(1):112. doi: 10.3390/ijms20010112. Int J Mol Sci. 2018. PMID: 30597961 Free PMC article. - The Role of Nrf2 Activity in Cancer Development and Progression.
Zimta AA, Cenariu D, Irimie A, Magdo L, Nabavi SM, Atanasov AG, Berindan-Neagoe I. Zimta AA, et al. Cancers (Basel). 2019 Nov 8;11(11):1755. doi: 10.3390/cancers11111755. Cancers (Basel). 2019. PMID: 31717324 Free PMC article. Review. - SIRT5 Promotes Cisplatin Resistance in Ovarian Cancer by Suppressing DNA Damage in a ROS-Dependent Manner via Regulation of the Nrf2/HO-1 Pathway.
Sun X, Wang S, Gai J, Guan J, Li J, Li Y, Zhao J, Zhao C, Fu L, Li Q. Sun X, et al. Front Oncol. 2019 Aug 13;9:754. doi: 10.3389/fonc.2019.00754. eCollection 2019. Front Oncol. 2019. PMID: 31456942 Free PMC article.
References
- Bouwman P., Aly A., Escandell J.M., Pieterse M., Bartkova J., van der Gulden H., Hiddingh S., Thanasoula M., Kulkarni A., Yang Q., et al. 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17:688–695 10.1038/nsmb.1831 - DOI - PMC - PubMed
- Bunting S.F., Callén E., Wong N., Chen H.T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 141:243–254 10.1016/j.cell.2010.03.012 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous