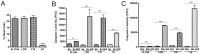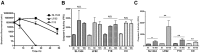Increased S-nitrosylation and proteasomal degradation of caspase-3 during infection contribute to the persistence of adherent invasive Escherichia coli (AIEC) in immune cells - PubMed (original) (raw)
Increased S-nitrosylation and proteasomal degradation of caspase-3 during infection contribute to the persistence of adherent invasive Escherichia coli (AIEC) in immune cells
Karl A Dunne et al. PLoS One. 2013.
Abstract
Adherent invasive Escherichia coli (AIEC) have been implicated as a causative agent of Crohn's disease (CD) due to their isolation from the intestines of CD sufferers and their ability to persist in macrophages inducing granulomas. The rapid intracellular multiplication of AIEC sets it apart from other enteric pathogens such as Salmonella Typhimurium which after limited replication induce programmed cell death (PCD). Understanding the response of infected cells to the increased AIEC bacterial load and associated metabolic stress may offer insights into AIEC pathogenesis and its association with CD. Here we show that AIEC persistence within macrophages and dendritic cells is facilitated by increased proteasomal degradation of caspase-3. In addition S-nitrosylation of pro- and active forms of caspase-3, which can inhibit the enzymes activity, is increased in AIEC infected macrophages. This S-nitrosylated caspase-3 was seen to accumulate upon inhibition of the proteasome indicating an additional role for S-nitrosylation in inducing caspase-3 degradation in a manner independent of ubiquitination. In addition to the autophagic genetic defects that are linked to CD, this delay in apoptosis mediated in AIEC infected cells through increased degradation of caspase-3, may be an essential factor in its prolonged persistence in CD patients.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
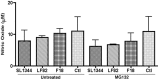
Figure 1. Caspase-3 accumulates in LF82 infected RAW 264.7 cells after proteasome inhibition.
(A) Cell death of RAW 264.7 cells at 6 hours post infection (hpi). The percentage of dead cells were calculated using a LIVE/DEAD stain and expressed as a percentage of the total number of cells present per well of a 24 well plate. (B) Accumulation of caspase-3 activity in RAW 264.7 cells after proteasome inhibition from 0–6 hpi. Caspase-3 activity in RAW 264.7 cells was measured at 6 hpi and expressed as activity, measured in fluorescence focus units (FFU) per mg of protein. Measurements were carried out in the presence or absence of 10 µM MG132. (C) Accumulation of caspase-3 activity in RAW 264.7 cells after proteasome inhibition from 24–30 hpi. Caspase-3 activity in RAW 264.7 cells was measured at 30 hpi and expressed as activity, measured in fluorescence focus units (FFU), per mg of protein. Measurements were carried out in the presence or absence of 10 µM MG132. All experiments were repeated at least three times in triplicate and data from representative experiments are shown. Data was analyzed by an unpaired Student’s _t_-test. Statistically significant relationships are denoted. P values **<0.05, ***<0.005.
Figure 2. Caspase-3 accumulates post-proteasome inhibition in LF82 BMDCs.
(A) Recovery of bacteria post intracellular survival and growth in BMDCs. After infection of BMDCs at an MOI of 100, bacteria were recovered and counted over 48 hpi. Intracellular growth curves were repeated at least three times in triplicate and data from a representative experiment is shown. (B) Caspase-3 activity in BMDCs at 3, 6 and 10 hpi. Caspase-3 activity was measured for the first 10 hpi and expressed as activity per mg of protein recovered. Caspase-3 activity assays were repeated at least three times in triplicate and data from a representative experiment is shown. No significant difference was noted between LF82 infected and any of the other infected or uninfected control cells. (C) Accumulation of caspase-3 activity in BMDCs after proteasome inhibition from 0–6 hpi. Caspase-3 activity in BMDCs was measured at 6 hpi and expressed as activity, measured in fluorescence focus units (FFU) per mg of protein. Measurements were carried out in the presence or absence of 10 µM MG132. All experiments were repeated at least three times in triplicate and data from a representative experiment is shown. Data was analyzed by an unpaired Student’s _t_-test. Statistically significant relationships are denoted. NS = Not significant. P values *<0.01, **<0.05.
Figure 3. The effect of proteasome inhibition on caspase-3 activity in T84 intestinal epithelial cells from 0–6 hpi.
Caspase-3 activity in T84s was measured at 6 hpi and expressed as activity, measured in fluorescence focus units (FFU) per mg of protein. Measurements were carried out in the presence or absence of 10 µM MG132. Experiments were repeated at least three times in triplicate and data from a representative experiment is shown. Data was analyzed by an unpaired Student’s _t_-test. NS = Not significant.
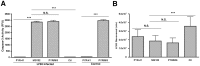
Figure 4. Caspase-3 build up in RAW 264.7 cells post-proteasome inhibition is independent of ubiquitination.
(A) The effect of inhibition of E1 ubiquitin activating enzymes on caspase-3 accumulation. RAW 264.7 cells were infected with LF82 or left uninfected (control) and treated for 6 hpi with 10 µM MG132, 50 µM PYR-41 or a combination thereof (PYR/MG). Caspase-3 activity was measured and expressed as activity per mg of protein recovered. Infected but untreated RAW 264.7 cells were used as controls also. Measurements were carried out in the presence or absence of 10 µM MG132. (B) The effect of inhibition of ubiquitination and proteasomal degradation on bacterial intracellular survival. RAW 264.7 cells were infected with LF82 and treated for 6 hpi with 10 µM MG132, 50 µM PYR-41 or a combination thereof (PYR/MG). Bacterial colony counts were then carried out on bacteria recovered from each well and expressed as colony forming units (cfu). Untreated and infected RAW 264.7 cells were used as an additional control. Experiments were repeated at least three times in triplicate and data from a representative experiment is shown. Data was analyzed by an unpaired Student’s _t_-test. Statistically significant relationships are denoted. NS = Not significant. P value **<0.05, ***<0.005.
Figure 5. Build-up of S-nitrosylated forms of caspase-3 in RAW 264.7 macrophages when the proteasome is inhibited.
S-nitrosylated proteins were probed with anti-caspase-3 antibody to examine accumulation of pro- (p32) and active (p17) forms of caspase-3 after inhibition of proteasomal degradation with 10 µM MG132. Western blotting was repeated three times and a representative Western blot is shown.
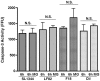
Figure 6. Nitric oxide production in RAW 264.7 cells 6 hpi.
NO levels in the supernatants of RAW 264.7 cells were measured using the Griess assay. Levels in cells treated with MG132 (10 µM) were lower than untreated cells but there was no significant difference between any of the uninfected samples or controls. This decrease in NO production upon MG132 addition indicated that the increase in S-nitrosylation of caspase-3 was not due to increased NO production in response to infection. Experiments were repeated at least three times in triplicate and data from a representative experiment is shown.
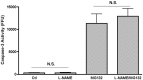
Figure 7. The effect of iNOS inhibition on the observed increase in caspase-3 activity post-proteasomal blocking.
RAW 264.7 cells were infected with LF82 and treated with or without the proteasomal inhibitor MG132 (10 µM) as before. Addition of the iNOS inhibitor L-NAME (100 µM) did not cause any decrease in caspase-3 activity in cells where the proteasome was blocked. _P_-value NS (Not significant).
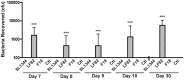
Figure 8. Recovery of bacteria from RAW 264.7 cells up to 30 days post infection.
RAW 264.7 cells were infected as described and maintained in 3% FCS for a period of 30 days with regular media changes and maintenance of gentamycin at 50 µg/ml. Intracellular bacteria were harvested as before by lysing RAW 264.7 cells. Experiments were repeated at least three times in triplicate and data from a representative experiment is shown. Data was analyzed by an unpaired Student’s _t_-test. Statistically significant relationships are denoted. P value ***<0.005.
Similar articles
- Crohn's disease-associated AIEC inhibiting intestinal epithelial cell-derived exosomal let-7b expression regulates macrophage polarization to exacerbate intestinal fibrosis.
Xu Y, Qian W, Huang L, Wen W, Li Y, Guo F, Zhu Z, Li Z, Gong J, Yu Z, Zhou Y, Lu N, Zhu W, Guo Z. Xu Y, et al. Gut Microbes. 2023 Jan-Dec;15(1):2193115. doi: 10.1080/19490976.2023.2193115. Gut Microbes. 2023. PMID: 36945126 Free PMC article. - Exosomes transfer miRNAs from cell-to-cell to inhibit autophagy during infection with Crohn's disease-associated adherent-invasive E. coli.
Larabi A, Dalmasso G, Delmas J, Barnich N, Nguyen HTT. Larabi A, et al. Gut Microbes. 2020 Nov 1;11(6):1677-1694. doi: 10.1080/19490976.2020.1771985. Epub 2020 Jun 25. Gut Microbes. 2020. PMID: 32583714 Free PMC article. - Development of Heptylmannoside-Based Glycoconjugate Antiadhesive Compounds against Adherent-Invasive Escherichia coli Bacteria Associated with Crohn's Disease.
Sivignon A, Yan X, Alvarez Dorta D, Bonnet R, Bouckaert J, Fleury E, Bernard J, Gouin SG, Darfeuille-Michaud A, Barnich N. Sivignon A, et al. mBio. 2015 Nov 17;6(6):e01298-15. doi: 10.1128/mBio.01298-15. mBio. 2015. PMID: 26578673 Free PMC article. - Adherent-invasive Escherichia coli in inflammatory bowel disease.
Rolhion N, Darfeuille-Michaud A. Rolhion N, et al. Inflamm Bowel Dis. 2007 Oct;13(10):1277-83. doi: 10.1002/ibd.20176. Inflamm Bowel Dis. 2007. PMID: 17476674 Review. - Adherent-invasive Escherichia coli: a putative new E. coli pathotype associated with Crohn's disease.
Darfeuille-Michaud A. Darfeuille-Michaud A. Int J Med Microbiol. 2002 Sep;292(3-4):185-93. doi: 10.1078/1438-4221-00201. Int J Med Microbiol. 2002. PMID: 12398209 Review.
Cited by
- Understanding host-adherent-invasive Escherichia coli interaction in Crohn's disease: opening up new therapeutic strategies.
Agus A, Massier S, Darfeuille-Michaud A, Billard E, Barnich N. Agus A, et al. Biomed Res Int. 2014;2014:567929. doi: 10.1155/2014/567929. Epub 2014 Dec 15. Biomed Res Int. 2014. PMID: 25580435 Free PMC article. Review. - Bacterial secreted effectors and caspase-3 interactions.
Wall DM, McCormick BA. Wall DM, et al. Cell Microbiol. 2014 Dec;16(12):1746-56. doi: 10.1111/cmi.12368. Epub 2014 Oct 30. Cell Microbiol. 2014. PMID: 25262664 Free PMC article. Review. - Crohn's Disease Pathobiont Adherent-Invasive E coli Disrupts Epithelial Mitochondrial Networks With Implications for Gut Permeability.
Mancini NL, Rajeev S, Jayme TS, Wang A, Keita ÅV, Workentine ML, Hamed S, Söderholm JD, Lopes F, Shutt TE, Shearer J, McKay DM. Mancini NL, et al. Cell Mol Gastroenterol Hepatol. 2021;11(2):551-571. doi: 10.1016/j.jcmgh.2020.09.013. Epub 2020 Sep 28. Cell Mol Gastroenterol Hepatol. 2021. PMID: 32992049 Free PMC article. - Inflammation associated ethanolamine facilitates infection by Crohn's disease-linked adherent-invasive Escherichia coli.
Ormsby MJ, Logan M, Johnson SA, McIntosh A, Fallata G, Papadopoulou R, Papachristou E, Hold GL, Hansen R, Ijaz UZ, Russell RK, Gerasimidis K, Wall DM. Ormsby MJ, et al. EBioMedicine. 2019 May;43:325-332. doi: 10.1016/j.ebiom.2019.03.071. Epub 2019 Apr 26. EBioMedicine. 2019. PMID: 31036531 Free PMC article. - Endothelial Nitric Oxide Synthase-Derived Nitric Oxide Prevents Dihydrofolate Reductase Degradation via Promoting S-Nitrosylation.
Cai Z, Lu Q, Ding Y, Wang Q, Xiao L, Song P, Zou MH. Cai Z, et al. Arterioscler Thromb Vasc Biol. 2015 Nov;35(11):2366-73. doi: 10.1161/ATVBAHA.115.305796. Epub 2015 Sep 17. Arterioscler Thromb Vasc Biol. 2015. PMID: 26381869 Free PMC article.
References
- Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, et al. (2004) High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127: 412–421. - PubMed
- Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, et al. (2007) Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J 1: 403–418. - PubMed
- Sasaki M, Sitaraman SV, Babbin BA, Gerner-Smidt P, Ribot EM, et al. (2007) Invasive Escherichia coli are a feature of Crohn’s disease. Lab Invest 87: 1042–1054. - PubMed
- Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, et al. (2004) Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology 127: 80–93. - PubMed
- Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, et al. (1998) Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology 115: 1405–1413. - PubMed
MeSH terms
Substances
Grants and funding
This work was supported by a Tenovus-Scotland Pump-priming award (http://www.tenovus-scotland.org.uk/) and by the BBSRC - Biotechnology and Biological Sciences Research Council Grant number BB/K008005/1. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials