Type III restriction-modification enzymes: a historical perspective - PubMed (original) (raw)
Review
Type III restriction-modification enzymes: a historical perspective
Desirazu N Rao et al. Nucleic Acids Res. 2014 Jan.
Abstract
Restriction endonucleases interact with DNA at specific sites leading to cleavage of DNA. Bacterial DNA is protected from restriction endonuclease cleavage by modifying the DNA using a DNA methyltransferase. Based on their molecular structure, sequence recognition, cleavage position and cofactor requirements, restriction-modification (R-M) systems are classified into four groups. Type III R-M enzymes need to interact with two separate unmethylated DNA sequences in inversely repeated head-to-head orientations for efficient cleavage to occur at a defined location (25-27 bp downstream of one of the recognition sites). Like the Type I R-M enzymes, Type III R-M enzymes possess a sequence-specific ATPase activity for DNA cleavage. ATP hydrolysis is required for the long-distance communication between the sites before cleavage. Different models, based on 1D diffusion and/or 3D-DNA looping, exist to explain how the long-distance interaction between the two recognition sites takes place. Type III R-M systems are found in most sequenced bacteria. Genome sequencing of many pathogenic bacteria also shows the presence of a number of phase-variable Type III R-M systems, which play a role in virulence. A growing number of these enzymes are being subjected to biochemical and genetic studies, which, when combined with ongoing structural analyses, promise to provide details for mechanisms of DNA recognition and catalysis.
Figures
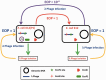
Figure 1.
Schematic illustration showing the presence of an R–M system in P1 prophage. This cartoon depicts the infection of λ phage isolated from E. coli K12 strain into E. coli K12(P1) lysogens and vice versa. The infection of an E. coli K12(P1) lysogen with λ phage isolated from E. coli K12 resulted in reduced e.o.p. However, infection of E. coli K12 with λ phage isolated from the E. coli K12(P1) lysogen was efficient. These experiments suggested that P1 phage harbors an independent R–M system, later identified as EcoP1.
Similar articles
- DNA cleavage by type III restriction-modification enzyme EcoP15I is independent of spacer distance between two head to head oriented recognition sites.
Mücke M, Reich S, Möncke-Buchner E, Reuter M, Krüger DH. Mücke M, et al. J Mol Biol. 2001 Sep 28;312(4):687-98. doi: 10.1006/jmbi.2001.4998. J Mol Biol. 2001. PMID: 11575924 - Mechanistic insights into type III restriction enzymes.
Raghavendra NK, Bheemanaik S, Rao DN. Raghavendra NK, et al. Front Biosci (Landmark Ed). 2012 Jan 1;17(3):1094-107. doi: 10.2741/3975. Front Biosci (Landmark Ed). 2012. PMID: 22201792 Review. - Type III restriction endonucleases translocate DNA in a reaction driven by recognition site-specific ATP hydrolysis.
Meisel A, Mackeldanz P, Bickle TA, Krüger DH, Schroeder C. Meisel A, et al. EMBO J. 1995 Jun 15;14(12):2958-66. doi: 10.1002/j.1460-2075.1995.tb07296.x. EMBO J. 1995. PMID: 7796821 Free PMC article. - Single-site DNA cleavage by Type III restriction endonuclease requires a site-bound enzyme and a trans-acting enzyme that are ATPase-activated.
Ahmad I, Kulkarni M, Gopinath A, Saikrishnan K. Ahmad I, et al. Nucleic Acids Res. 2018 Jul 6;46(12):6229-6237. doi: 10.1093/nar/gky344. Nucleic Acids Res. 2018. PMID: 29846668 Free PMC article. - Complex restriction enzymes: NTP-driven molecular motors.
Bourniquel AA, Bickle TA. Bourniquel AA, et al. Biochimie. 2002 Nov;84(11):1047-59. doi: 10.1016/s0300-9084(02)00020-2. Biochimie. 2002. PMID: 12595133 Review.
Cited by
- Is the Evolution of Salmonella enterica subsp. enterica Linked to Restriction-Modification Systems?
Roer L, Hendriksen RS, Leekitcharoenphon P, Lukjancenko O, Kaas RS, Hasman H, Aarestrup FM. Roer L, et al. mSystems. 2016 Jun 21;1(3):e00009-16. doi: 10.1128/mSystems.00009-16. eCollection 2016 May-Jun. mSystems. 2016. PMID: 27822532 Free PMC article. - Deciphering bacterial epigenomes using modern sequencing technologies.
Beaulaurier J, Schadt EE, Fang G. Beaulaurier J, et al. Nat Rev Genet. 2019 Mar;20(3):157-172. doi: 10.1038/s41576-018-0081-3. Nat Rev Genet. 2019. PMID: 30546107 Free PMC article. Review. - Streptococcus suis contains multiple phase-variable methyltransferases that show a discrete lineage distribution.
Atack JM, Weinert LA, Tucker AW, Husna AU, Wileman TM, F Hadjirin N, Hoa NT, Parkhill J, Maskell DJ, Blackall PJ, Jennings MP. Atack JM, et al. Nucleic Acids Res. 2018 Nov 30;46(21):11466-11476. doi: 10.1093/nar/gky913. Nucleic Acids Res. 2018. PMID: 30304532 Free PMC article. - Adenine methylation in eukaryotes: Apprehending the complex evolutionary history and functional potential of an epigenetic modification.
Iyer LM, Zhang D, Aravind L. Iyer LM, et al. Bioessays. 2016 Jan;38(1):27-40. doi: 10.1002/bies.201500104. Epub 2015 Dec 12. Bioessays. 2016. PMID: 26660621 Free PMC article. Review. - Improving transformation of Staphylococcus aureus belonging to the CC1, CC5 and CC8 clonal complexes.
Jones MJ, Donegan NP, Mikheyeva IV, Cheung AL. Jones MJ, et al. PLoS One. 2015 Mar 25;10(3):e0119487. doi: 10.1371/journal.pone.0119487. eCollection 2015. PLoS One. 2015. PMID: 25807379 Free PMC article.
References
- Arber W, Dussoix D. Host specificity of DNA produced by Escherichia coli. I. Host controlled modification of bacteriophage lambda. J. Mol. Biol. 1962;5:18–36. - PubMed
- Arber W. Host-controlled modification of bacteriophage. Annu. Rev .Microbiol. 1965;19:365–378. - PubMed
- Stacey KA. Intracellular modification of nucleic acids. Br. Med. Bull. 1965;21:211–216. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases