Translating dosage compensation to trisomy 21 - PubMed (original) (raw)
. 2013 Aug 15;500(7462):296-300.
doi: 10.1038/nature12394. Epub 2013 Jul 17.
Yuanchun Jing, Gregory J Cost, Jen-Chieh Chiang, Heather J Kolpa, Allison M Cotton, Dawn M Carone, Benjamin R Carone, David A Shivak, Dmitry Y Guschin, Jocelynn R Pearl, Edward J Rebar, Meg Byron, Philip D Gregory, Carolyn J Brown, Fyodor D Urnov, Lisa L Hall, Jeanne B Lawrence
Affiliations
- PMID: 23863942
- PMCID: PMC3848249
- DOI: 10.1038/nature12394
Translating dosage compensation to trisomy 21
Jun Jiang et al. Nature. 2013.
Abstract
Down's syndrome is a common disorder with enormous medical and social costs, caused by trisomy for chromosome 21. We tested the concept that gene imbalance across an extra chromosome can be de facto corrected by manipulating a single gene, XIST (the X-inactivation gene). Using genome editing with zinc finger nucleases, we inserted a large, inducible XIST transgene into the DYRK1A locus on chromosome 21, in Down's syndrome pluripotent stem cells. The XIST non-coding RNA coats chromosome 21 and triggers stable heterochromatin modifications, chromosome-wide transcriptional silencing and DNA methylation to form a 'chromosome 21 Barr body'. This provides a model to study human chromosome inactivation and creates a system to investigate genomic expression changes and cellular pathologies of trisomy 21, free from genetic and epigenetic noise. Notably, deficits in proliferation and neural rosette formation are rapidly reversed upon silencing one chromosome 21. Successful trisomy silencing in vitro also surmounts the major first step towards potential development of 'chromosome therapy'.
Conflict of interest statement
Conflict of interest statement. JBL and LLH are the inventors on an issued patent describing the concept of epigenetic chromosome therapy by targeted addition of non-coding RNA. GJC, DAS, DYG, JRP, EJR, PDG, FDU are full-time employees of Sangamo BioSciences.
Figures
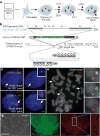
Figure 1. Genome-editing integrates XIST into Chr21 in trisomic iPSCs
a. Concept for translating dosage compensation to trisomy 21. b. XIST construct (19kb): two homologous arms and 14kb XIST cDNA with inducible pTRE3G promoter. c. DNA/RNA FISH in interphase DS iPSCs shows XIST overlaps one of three DYRK1A genes in non-expressing cell (top, arrows), and a large XIST RNA territory with DYRK1A after 3 days in dox (bottom, arrows). Scale: 2 μm. d. OCT4 immunostaining & XIST RNA FISH in a transgenic colony: highly consistent XIST expression throughout the colony. Scale: 100 μm. e. Metaphase DNA FISH shows one targeted Chr21. XIST gene (asterisk and close-up) overlaps one of three DYRK1A genes (arrows).
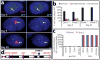
Figure 2. XIST induces heterochromatin modifications and condensed Chr21 Barr Body
a. XIST RNA recruits heterochromatic epigenetic marks (e.g. UbH2A). Channels separated for indicated cell. b. Percentage of XIST territories with heterochromatin marks. Mean ± SE, 100 nuclei in ~5 colonies. c. XIST RNA induces “Chr21 Barr Body” visible by DAPI stain (arrow). Scale: 2 μm.
Figure 3. XIST induces long-range silencing in targeted iPSCs
a. RNA FISH. APP RNA transcribes from three loci in uninduced cells (Day 0), and is progressively silenced following induction (targeted Chr21, arrows). Scale: 2 μm. b. Quantification of APP silencing. Mean ± SE, 100 nuclei. c. Silencing for four more Chr21-linked genes by RNA FISH. Mean ± SE from 100 nuclei. d. Long range silencing of Chr21 genes by XIST RNA. USP25 is ~21 Mb from XIST integration site (black arrow).
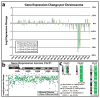
Figure 4. Genomic expression and methylation reveal widespread silencing of Chr21
a. Microarray: expression difference for three transgenic clones in dox versus no dox, compared to disomic line versus trisomic parental line. Total change in gene expression (N=3) per chromosome shows Chr21 “correction” near disomic levels, with only limited changes on other chromosomes. Right Y-axis scaled for percent gene expression change. b. Distribution of individual gene repression across Chr21. c. Methylation of CpG island promoters. In treated clones, 97% of Chr21 genes increased by at least 5% (2 fold greater than average), compared to none in the parental line. P: parental line; 1: Clone 1; 2: Clone 2.
Figure 5. “Trisomy correction” effects cell proliferation and neurogenesis
a. One week of XIST expression resulted in larger, more numerous colonies (representative sample). b. Changes in cell number for parental line (PL), parental line subclone (PL-s), and six transgenic clones (C1–C6). Mean ±SE. (n=4–6). c. Corrected cultures formed neural rosettes by day 14; trisomic (parental and non-induced) cultures took longer (17days). Scale: 100μm. e. Number of rosettes formed on day 14 and day 17. Mean ±SE, 10–12 random fields in triplicate. P: parental; C1: Clone 1; C3: Clone 3.
Comment in
- How to correct chromosomal trisomy.
Disteche CM. Disteche CM. Cell Res. 2013 Dec;23(12):1345-6. doi: 10.1038/cr.2013.135. Epub 2013 Oct 1. Cell Res. 2013. PMID: 24080727 Free PMC article.
Similar articles
- Silencing Trisomy 21 with XIST in Neural Stem Cells Promotes Neuronal Differentiation.
Czermiński JT, Lawrence JB. Czermiński JT, et al. Dev Cell. 2020 Feb 10;52(3):294-308.e3. doi: 10.1016/j.devcel.2019.12.015. Epub 2020 Jan 23. Dev Cell. 2020. PMID: 31978324 Free PMC article. - How to correct chromosomal trisomy.
Disteche CM. Disteche CM. Cell Res. 2013 Dec;23(12):1345-6. doi: 10.1038/cr.2013.135. Epub 2013 Oct 1. Cell Res. 2013. PMID: 24080727 Free PMC article. - Trisomy silencing by XIST normalizes Down syndrome cell pathogenesis demonstrated for hematopoietic defects in vitro.
Chiang JC, Jiang J, Newburger PE, Lawrence JB. Chiang JC, et al. Nat Commun. 2018 Dec 5;9(1):5180. doi: 10.1038/s41467-018-07630-y. Nat Commun. 2018. PMID: 30518921 Free PMC article. - Trisomy silencing by XIST: translational prospects and challenges.
Gupta K, Czerminski JT, Lawrence JB. Gupta K, et al. Hum Genet. 2024 Jul;143(7):843-855. doi: 10.1007/s00439-024-02651-8. Epub 2024 Mar 9. Hum Genet. 2024. PMID: 38459355 Free PMC article. Review. - The making of a Barr body: the mosaic of factors that eXIST on the mammalian inactive X chromosome.
Dixon-McDougall T, Brown C. Dixon-McDougall T, et al. Biochem Cell Biol. 2016 Feb;94(1):56-70. doi: 10.1139/bcb-2015-0016. Epub 2015 Jun 24. Biochem Cell Biol. 2016. PMID: 26283003 Review.
Cited by
- Concise Review: Methods and Cell Types Used to Generate Down Syndrome Induced Pluripotent Stem Cells.
Hibaoui Y, Feki A. Hibaoui Y, et al. J Clin Med. 2015 Apr 15;4(4):696-714. doi: 10.3390/jcm4040696. J Clin Med. 2015. PMID: 26239351 Free PMC article. Review. - An Integrated Human/Murine Transcriptome and Pathway Approach To Identify Prenatal Treatments For Down Syndrome.
Guedj F, Pennings JL, Massingham LJ, Wick HC, Siegel AE, Tantravahi U, Bianchi DW. Guedj F, et al. Sci Rep. 2016 Sep 2;6:32353. doi: 10.1038/srep32353. Sci Rep. 2016. PMID: 27586445 Free PMC article. - Co-expression network analysis of Down's syndrome based on microarray data.
Zhao J, Zhang Z, Ren S, Zong Y, Kong X. Zhao J, et al. Exp Ther Med. 2016 Sep;12(3):1503-1508. doi: 10.3892/etm.2016.3462. Epub 2016 Jun 17. Exp Ther Med. 2016. PMID: 27588071 Free PMC article. - Loss of XIST in Breast Cancer Activates MSN-c-Met and Reprograms Microglia via Exosomal miRNA to Promote Brain Metastasis.
Xing F, Liu Y, Wu SY, Wu K, Sharma S, Mo YY, Feng J, Sanders S, Jin G, Singh R, Vidi PA, Tyagi A, Chan MD, Ruiz J, Debinski W, Pasche BC, Lo HW, Metheny-Barlow LJ, D'Agostino RB Jr, Watabe K. Xing F, et al. Cancer Res. 2018 Aug 1;78(15):4316-4330. doi: 10.1158/0008-5472.CAN-18-1102. Epub 2018 Jul 19. Cancer Res. 2018. PMID: 30026327 Free PMC article. - Zinc finger nuclease-mediated targeting of multiple transgenes to an endogenous soybean genomic locus via non-homologous end joining.
Bonawitz ND, Ainley WM, Itaya A, Chennareddy SR, Cicak T, Effinger K, Jiang K, Mall TK, Marri PR, Samuel JP, Sardesai N, Simpson M, Folkerts O, Sarria R, Webb SR, Gonzalez DO, Simmonds DH, Pareddy DR. Bonawitz ND, et al. Plant Biotechnol J. 2019 Apr;17(4):750-761. doi: 10.1111/pbi.13012. Epub 2018 Oct 15. Plant Biotechnol J. 2019. PMID: 30220095 Free PMC article.
References
- Megarbane A, et al. The 50th anniversary of the discovery of trisomy 21: the past, present, and future of research and treatment of Down syndrome. Genet Med. 2009;11:611–616. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM085548/GM/NIGMS NIH HHS/United States
- GM085548/GM/NIGMS NIH HHS/United States
- F32 CA154086/CA/NCI NIH HHS/United States
- 1F32CA154086/CA/NCI NIH HHS/United States
- GM053234/GM/NIGMS NIH HHS/United States
- T32 HD007439/HD/NICHD NIH HHS/United States
- R01 GM053234/GM/NIGMS NIH HHS/United States
- RC4 GM096400/GM/NIGMS NIH HHS/United States
- MOP-13680/CAPMC/ CIHR/Canada
- 2T32HD007439/HD/NICHD NIH HHS/United States
- GM096400 RC4/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials