DNA in motion during double-strand break repair - PubMed (original) (raw)
Review
DNA in motion during double-strand break repair
Judith Miné-Hattab et al. Trends Cell Biol. 2013 Nov.
Abstract
DNA organization and dynamics profoundly affect many biological processes such as gene regulation and DNA repair. In this review, we present the latest studies on DNA mobility in the context of DNA damage. Recent studies demonstrate that DNA mobility is dramatically increased in the presence of double-strand breaks (DSBs) in the yeast Saccharomyces cerevisiae. As a consequence, chromosomes explore a larger nuclear volume, facilitating homologous pairing but also increasing the rate of ectopic recombination. Increased DNA dynamics is dependent on several homologous recombination (HR) proteins and we are just beginning to understand how chromosome dynamics is regulated after DNA damage.
Keywords: DNA mobility; DNA repair; double-strand break repair; homologous recombination.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
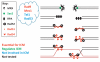
Figure 1
Depiction of the different homologous recombination (HR) proteins and their role in increasing the local mobility of the damaged site. Red: recombination proteins that are essential for increased chromosome mobility (ICM). Green: recombination proteins that regulate ICM by changing its timing. Black: recombination proteins that are not involved in ICM. Grey: recombination proteins that have not yet been tested. It is unknown precisely at which steps the proteins in the ‘cloud’ act.
Figure 2
Examples of the dynamics of URA3 loci (chromosome V) in budding yeast as a function of the number of double-strand breaks (DSBs) in the nucleus. The lines indicate a 2D projection of the trajectories of the two_URA3_ loci taken at between 10- and 30-s time intervals for approximately 15 min. (A) In the absence of DSBs, the two homologous loci are distant and explore only 3% of the nuclear volume.(B) In the presence of one to four DSBs induced on chromosomes III, the mobility of the URA3 loci increases and each locus can explore 11% of the nuclear volume. (C) After about 20 random γ-irradiation-induced DSBs per nucleus (200 Gy), _URA3_loci explore almost the entire nuclear volume and their trajectories overlap. The scale is 1 μm. These three examples illustrate that more DSBs in the nucleus induce greater DNA mobility, thereby increasing the probability of collisions between loci. With permission from [72].
Figure 3
The ‘collision’ and ‘altered chromatin’ models for increased chromosome mobility (ICM). A cut view of the nucleus is depicted. Dark blue represents the general chromatin state of one chromosome and light blue that of another. The mobility of a single intact locus on the light blue chromosome is colored red and the surrounding waves represent its mobility (from one to three, which reflect different confinement radii). Before a double-strand break (DSB), there is limited mobility (one wave on each side of the red focus). After a DSB, the mobility of the broken locus increases locally (three yellow waves on each side). In the collision model, the mobility of the intact red locus increases (shown as two red waves on each side of the locus) due to collisions with the broken locus provoking global ICM. In the altered chromatin model, the state of the chromatin, depicted as less entangled, changes globally as a result of DNA damage. This change in state increases the mobility of the whole genome (represented by the two waves surrounding the red locus) independently of the local ICM at the damaged locus (three yellow waves on each side).
Similar articles
- Poetry in motion: Increased chromosomal mobility after DNA damage.
Smith MJ, Rothstein R. Smith MJ, et al. DNA Repair (Amst). 2017 Aug;56:102-108. doi: 10.1016/j.dnarep.2017.06.012. Epub 2017 Jun 9. DNA Repair (Amst). 2017. PMID: 28663070 Free PMC article. Review. - Asymmetric Processing of DNA Ends at a Double-Strand Break Leads to Unconstrained Dynamics and Ectopic Translocation.
Marcomini I, Shimada K, Delgoshaie N, Yamamoto I, Seeber A, Cheblal A, Horigome C, Naumann U, Gasser SM. Marcomini I, et al. Cell Rep. 2018 Sep 4;24(10):2614-2628.e4. doi: 10.1016/j.celrep.2018.07.102. Cell Rep. 2018. PMID: 30184497 - Chromatin mobility upon DNA damage: state of the art and remaining questions.
Zimmer C, Fabre E. Zimmer C, et al. Curr Genet. 2019 Feb;65(1):1-9. doi: 10.1007/s00294-018-0852-6. Epub 2018 Jun 8. Curr Genet. 2019. PMID: 29947969 Review. - Pathways and assays for DNA double-strand break repair by homologous recombination.
Li J, Sun H, Huang Y, Wang Y, Liu Y, Chen X. Li J, et al. Acta Biochim Biophys Sin (Shanghai). 2019 Sep 6;51(9):879-889. doi: 10.1093/abbs/gmz076. Acta Biochim Biophys Sin (Shanghai). 2019. PMID: 31294447 Review. - Measuring Chromosome Pairing During Homologous Recombination in Yeast.
Joseph F, Lee SJ, Bryant EE, Rothstein R. Joseph F, et al. Methods Mol Biol. 2021;2153:253-265. doi: 10.1007/978-1-0716-0644-5_18. Methods Mol Biol. 2021. PMID: 32840785
Cited by
- Complex Chromatin Motions for DNA Repair.
Miné-Hattab J, Chiolo I. Miné-Hattab J, et al. Front Genet. 2020 Aug 27;11:800. doi: 10.3389/fgene.2020.00800. eCollection 2020. Front Genet. 2020. PMID: 33061931 Free PMC article. Review. - Multi-scale tracking reveals scale-dependent chromatin dynamics after DNA damage.
Miné-Hattab J, Recamier V, Izeddin I, Rothstein R, Darzacq X. Miné-Hattab J, et al. Mol Biol Cell. 2017 Aug 9;28(23):3323-32. doi: 10.1091/mbc.E17-05-0317. Online ahead of print. Mol Biol Cell. 2017. PMID: 28794266 Free PMC article. - Double-strand break repair and mis-repair in 3D.
Zagelbaum J, Gautier J. Zagelbaum J, et al. DNA Repair (Amst). 2023 Jan;121:103430. doi: 10.1016/j.dnarep.2022.103430. Epub 2022 Nov 17. DNA Repair (Amst). 2023. PMID: 36436496 Free PMC article. - RecA: Regulation and Mechanism of a Molecular Search Engine.
Bell JC, Kowalczykowski SC. Bell JC, et al. Trends Biochem Sci. 2016 Jun;41(6):491-507. doi: 10.1016/j.tibs.2016.04.002. Epub 2016 May 4. Trends Biochem Sci. 2016. PMID: 27156117 Free PMC article. Review. - The Knowns Unknowns: Exploring the Homologous Recombination Repair Pathway in Toxoplasma gondii.
Fenoy IM, Bogado SS, Contreras SM, Gottifredi V, Angel SO. Fenoy IM, et al. Front Microbiol. 2016 May 3;7:627. doi: 10.3389/fmicb.2016.00627. eCollection 2016. Front Microbiol. 2016. PMID: 27199954 Free PMC article. Review.
References
- Cremer C, et al. Detection of laser–UV microirradiation-induced DNA photolesions by immunofluorescent staining. Hum. Genet. 1980;54:107–110. - PubMed
- Marshall WF, et al. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 1997;7:930–939. - PubMed
- Heun P, et al. Chromosome dynamics in the yeast interphase nucleus. Science. 2001;294:2181–2186. - PubMed
- Berger AB, et al. High-resolution statistical mapping reveals gene territories in live yeast. Nat. Methods. 2008;5:1031–1037. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials