Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy - PubMed (original) (raw)
Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy
Adam I Fogel et al. Mol Cell Biol. 2013 Sep.
Abstract
During autophagy, a double membrane envelops cellular material for trafficking to the lysosome. Human beclin-1 and its yeast homologue, Atg6/Vps30, are scaffold proteins bound in a lipid kinase complex with multiple cellular functions, including autophagy. Several different Atg6 complexes exist, with an autophagy-specific form containing Atg14. However, the roles of Atg14 and beclin-1 in the activation of this complex remain unclear. We here addressed the mechanism of beclin-1 complex activation and reveal two critical steps in this pathway. First, we identified a unique domain in beclin-1, conserved in the yeast homologue Atg6, which is involved in membrane association and, unexpectedly, controls autophagosome size and number in yeast. Second, we demonstrated that human Atg14 is critical in controlling an autophagy-dependent phosphorylation of beclin-1. We map these novel phosphorylation sites to serines 90 and 93 and demonstrate that phosphorylation at these sites is necessary for maximal autophagy. These results help clarify the mechanism of beclin-1 and Atg14 during autophagy.
Figures
Fig 1
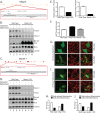
Knockout of hAtg14/Barkor and beclin-1 using TALE nuclease technology impairs autophagy. (A) Strategy for knocking out hAtg14 using transcription activator-like effector (TALE) nucleases. Exon 4 was targeted in the region shown, with the two TALE nuclease binding sequences indicated by underlining. Clones were selected based on HindIII digest after PCR of the region of interest. (B) Immunoblot analysis of hAtg14 knockout (KO) cells under normal and treated conditions. Cells were starved and/or treated with bafilomycin A1 (50 nM) for 3 h, and total lysates analyzed by immunoblotting for the proteins indicated. Ub, ubiquinated. (C) Strategy for knocking out beclin-1 using TALE nucleases. Exon 2 was targeted in the region shown, with the two TALE nuclease binding sequences indicated by underlining. Clones were selected based on PstI digest after PCR of the region of interest. (D) Immunoblot analysis of beclin-1 KO cells under normal and treated conditions. Cells were starved and/or treated with bafilomycin A1 (50 nM) for 3 h, and total lysates analyzed by immunoblotting for the proteins indicated. (E) Quantification of beclin-1 levels in hAtg14 KO cells and of hAtg14 levels in beclin-1 KO cells. Levels were normalized to wild-type protein levels found in HCT 116 cells. (F) Quantification of p62/SQSTM1 transcript levels, as assessed by TaqMan assay on triplicate samples from wild-type HCT 116, hAtg14 KO, or beclin-1 KO cells. (G) Analysis of mitophagy in wild-type or hAtg14 KO cells. Cells transfected with YFP-parkin were treated with vehicle or valinomycin at 10 μM for 24 h and analyzed for mitophagy by staining for Tom20. Significant mitochondrial clearance was observed in wild-type HCT 116 but not in hAtg14 KO cells, which mostly displayed trapped mitochondrial clusters. (H) Quantification of results of experiments for which representative images are shown in panel G. (I) Wild-type or beclin-1 KO cells transiently expressing YFP-parkin were treated with valinomycin at 10 μM for 24 h and analyzed for mitophagy by staining for Tom20. Beclin-1 KO cells showed a defect in clearing damaged mitochondria. (J) Quantification of results of experiments for which representative images are shown in panel I. Error bars show standard errors of the mean.
Fig 2
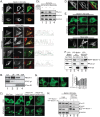
Association of beclin-1 with membranes involves the C-terminal domain. (A) Beclin-1 KO cells expressing YFP–beclin-1 and starved for 2 h were counterstained for mitochondria (Tom20) (top). HeLa cells expressing YFP–beclin-1 and counterstained for cytochrome c to mark mitochondria (Tom20), endoplasmic reticulum (KDEL), Golgi body (GM130), or early endosomes (EEA1) under starvation conditions (bottom). (B) HeLa cells were fractionated into cytosol (Cyt.), light membranes (LM), heavy membranes (HM), and then carbonate-extracted heavy membranes (ExM) and analyzed for endogenous beclin-1. PLC γ-1 and Tom20 serve as markers of the cytosolic and heavy membrane fractions, respectively. (Ci) YFP–beclin-1 was expressed ectopically in wild-type cells, hAtg14 KO cells, hAtg14 KO cells stably reexpressing hAtg14, or hAtg14 KO cells transfected with UVRAG siRNA and analyzed under fed and starved conditions. (Cii) Prior to transfection, immunoblotting for endogenous beclin-1 and UVRAG was performed on the samples used in the experiments for which representative images are shown in panel Ci, with immunoblotting for GAPDH serving as a loading control. (D) Membrane binding of YFP–beclin-1 still occurs in Bcl-2 binding-deficient beclin-1 F123A. HeLa cells expressing beclin-1 F123A were fixed and counterstained for the mitochondrial marker cytochrome c (Cyt C). (E) Confocal imaging of ectopically expressed YFP–beclin-1, YFP–beclin-1 lacking the C-terminal domain, or the YFP-tagged beclin-1 C-terminal domain alone (YFP 425–450) in beclin-1 KO cells under nutrient-rich or starvation conditions. Similar results were seen in HeLa cells. (F) Fractionation of cellular lysates from HeLa cells expressing the constructs used in the experiments whose results are shown in panel E was analyzed by immunoblotting. PLC γ-1 and Tim23 serve as markers of cytosolic and membrane fractions, respectively. (G) Alanine scanning mutagenesis of C-terminal large hydrophobic residues. YFP–beclin-1 W425A and YFP–beclin-1 ΔC had similar reductions in the degree of membrane association upon starvation. (H) Immunoprecipitation with GFP-Trap of beclin-1 and beclin-1 ΔC from cells co-overexpressing YFP–beclin-1 (or ΔC), Vps34-Flag, and Flag-Vps15.
Fig 3
The C-terminal domain (CTD) of beclin-1 is conserved in the yeast Atg6 and required for membrane association and cell survival in response to starvation. (A) Sequence alignment of the conserved CTD (orange) of human beclin-1 and yeast Atg6, in schematic with other known domains. (B) Confocal imaging of yeast cells expressing Atg6-GFP or Atg6 ΔC-GFP from the endogenous promoter and coexpressing plasmid-borne RFP-Cvt19 as a PAS marker. Colocalizing regions of GFP-Atg6 puncta with RFP-Cvt19 are indicated by arrowheads. No significant difference was observed upon nutrient deprivation (not shown). (C) _Atg6_Δ cells or cells expressing wild-type Atg6 or Atg6 ΔC were plated out and starved for the times indicated, and the cell survival rate quantified. Error bars show standard standard errors of the mean. Unpaired t test. * denotes P < 0.05.
Fig 4
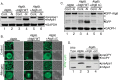
The CTD is required for specific (Cvt) autophagy, macroautophagy, and vacuolar protein sorting. (A) Analysis of Cvt pathway autophagy by aminopeptidase-I processing. _Atg6_Δ cells or those cells rescued with Atg6 or Atg6 ΔC were depleted of nitrogen (-N) for 3 h, and the cellular lysates analyzed by immunoblotting for Ape1. GAPDH serves as a loading control. (B) Analysis of autophagy in yeast using an Atg8-GFP strain. _Atg6_Δ cells, _Atg6_Δ cells expressing Atg6, or Atg6 ΔC cells were subjected to control or nitrogen-depleted conditions and analyzed by confocal microscopy. Scale bars are 5 μm. (C) Immunoblot for GFP-Atg8 processing in the same strains used in the experiments for which representative results are shown in panel B. GAPDH serves as a loading control. Open arrowheads denote background bands. (D) Wild-type cells, _Atg6_Δ cells, or _Atg6_Δ cells expressing Atg6-RFP (+Atg6) and Atg6 ΔC-RFP (+ΔC) from the genomic Atg6 locus were grown to mid-log phase and analyzed for their capacity to carry out vacuolar protein sorting, as assessed by carboxypeptidase Y (CPY) cleavage. Immunoblotting for Ape1 cleavage serves to control for defects in specific autophagy due to Atg6 deletion. preApe1, precursor Ape1; mApe1, mature Ape1; preCPY, precursor CPY; mCPY, mature CPY.
Fig 5
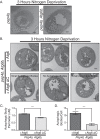
The Atg6 C-terminal domain controls autophagic body size and number. (A) _Pep4_Δ or _Pep4_Δ _Atg6_Δ cells were starved and analyzed by electron microscopy for autophagic body formation. (B) Electron microscopy analysis of _Pep4_Δ _Atg6_Δ cells expressing either wild-type Atg6 or Atg6 ΔC. _Pep4_Δ _Atg6_Δ cells expressing Atg6 had autophagic bodies morphologically similar to those in _Pep4_Δ cells, whereas cells expressing Atg6 ΔC had either small autophagic bodies or no autophagic bodies, as shown in the three sample images. (C) Quantification of autophagic body perimeters in cells used in the experiments for which representative images are shown in panel B. (D) Quantification of autophagic body numbers in cells used in the experiments for which representative images are shown in panel B. Scale bar is 500 nm in all images. Error bars show standard errors of the mean. Unpaired t test. *** denotes P < 0.0001.
Fig 6
Beclin-1 is phosphorylated during autophagy and mitophagy. (A) Phos-tag immunoblot analysis of beclin-1 during starvation. Wild-type HCT 116 cells were maintained in nutrient-rich medium (fed) or starved for 2 h (ST). Starved lysates were treated with calf intestinal phosphatase (ST+CIP) to remove phosphate groups and analyzed by immunoblotting after Phos-tag or regular SDS-PAGE. (B) Reversal of autophagic stimuli removes beclin-1 phosphorylation. Lysates were collected and analyzed after incubation in starvation medium for the indicated times or after 2 h under starvation conditions followed by a 2-h chase in normal growth medium (lane 6). (C) Beclin-1 phosphorylation occurs during mitochondrial autophagy. HCT 116 cells stably expressing YFP-parkin, an effector of mitochondrial autophagy, were treated with the mitochondrial uncoupler valinomycin (10 μM) for the indicated times, and the lysates analyzed by Phos-tag or normal SDS-PAGE, followed by immunoblotting for the indicated proteins.
Fig 7
Phosphorylation of serines 90 and 93 in beclin-1 contributes to autophagic function. (A) Beclin-1 KO cells or beclin-1 KO cells expressing beclin-1 wild type or indicated mutant were analyzed by immunoblotting after Phos-tag or regular SDS-PAGE. This approach identified serines 90 and 93 as responsible for the supershifting observed when performing Phos-tag immunoblotting of beclin-1 (lanes 8, 9, and 14). (B) Beclin-1 phosphorylation sites are conserved in higher eukaryotes. Sequence alignment of beclin-1 phosphorylation sites and surrounding amino acids of the indicated species. Phosphorylated serines are highlighted in green. (C) Beclin-1 KO cells were transfected with YFP–beclin-1 wild type or YFP–beclin-1 S90A/S93A, incubated under nutrient-rich or starvation conditions, and analyzed by confocal microscopy. (D) Complex formation in beclin-1 KO cells stably reexpressing wild-type or S90A/S93A YFP–beclin-1. Cell lysates were prepared, and complexes purified by incubation with GFP-Trap beads and analyzed by immunoblotting for the indicated proteins. Equal retention of binding partners was observed regardless of mutation. (E) Beclin-1 KO cells stably reexpressing wild-type, S90A/S93A, or S90E/S93E beclin-1 were compared with KO cells for the ability to turn over p62 in response to starvation. (F) p62 turnover in the cell lines used in the experiments for which results are shown in panel E was assessed by immunofluorescence detection of endogenous p62 under both nutrient-rich and starvation conditions; n = 3 separate experiments. (G) Quantification of results from experiments for which representative images are shown in panel F. Error bars show standard errors of the mean. AU, arbitrary units.
Fig 8
Beclin-1 phosphorylation is dependent on hAtg14. (A) Strategy for knocking out Atg5 using TALE nucleases. Exon 2 was targeted in the region shown, with the two TALE nuclease binding sequences indicated by underlining. Clones were selected based on EcoRI digest after PCR of the region of interest. (B) Wild-type HCT 116 cells, hAtg14 KO cells, hAtg14 KO cells stably reexpressing hAtg14, or Atg5 KO cells were incubated under nutrient-rich or starvation conditions for 2 h, and cellular lysates analyzed by both Phos-tag and regular SDS-PAGE. A long exposure of a Phos-tag immunoblot of beclin-1 serves to emphasize the very minimal level of beclin-1 phosphorylation in the absence of hAtg14. Immunoblotting for LC3 and GAPDH controlled for autophagy and protein loading, respectively. (C) Quantification of the Phos-tag analysis (nonsaturating exposure) shown in panel B. (D) Strategy for knocking out Atg13 using TALE nucleases. Exon 9 was targeted in the region shown, with the two TALE nuclease binding sequences indicated by underlining. Clones were selected based on PvuII digest after PCR of the region of interest. (E) Analysis of Atg13 KO cells for beclin-1 phosphorylation. Wild-type HCT 116 cells, hAtg14 KO cells, hAtg14 KO cells stably reexpressing hAtg14, or Atg13 KO cells were incubated under nutrient-rich or starvation conditions for 2 h, and cellular lysates were analyzed by both Phos-tag and regular SDS-PAGE. (F) Analysis of beclin-1 phosphorylation after UVRAG siRNA treatment. HCT 116 cells were transfected with siRNA against UVRAG and, after 2 days of expression, incubated under nutrient-rich or starvation conditions for 2 h, and cellular lysates were analyzed by both Phos-tag and regular SDS-PAGE. (G) Quantification of the Phos-tag analysis shown in panel F.
Similar articles
- Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG.
Itakura E, Kishi C, Inoue K, Mizushima N. Itakura E, et al. Mol Biol Cell. 2008 Dec;19(12):5360-72. doi: 10.1091/mbc.e08-01-0080. Epub 2008 Oct 8. Mol Biol Cell. 2008. PMID: 18843052 Free PMC article. - Nrbf2 protein suppresses autophagy by modulating Atg14L protein-containing Beclin 1-Vps34 complex architecture and reducing intracellular phosphatidylinositol-3 phosphate levels.
Zhong Y, Morris DH, Jin L, Patel MS, Karunakaran SK, Fu YJ, Matuszak EA, Weiss HL, Chait BT, Wang QJ. Zhong Y, et al. J Biol Chem. 2014 Sep 19;289(38):26021-26037. doi: 10.1074/jbc.M114.561134. Epub 2014 Aug 1. J Biol Chem. 2014. PMID: 25086043 Free PMC article. - Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L).
Fan W, Nassiri A, Zhong Q. Fan W, et al. Proc Natl Acad Sci U S A. 2011 May 10;108(19):7769-74. doi: 10.1073/pnas.1016472108. Epub 2011 Apr 25. Proc Natl Acad Sci U S A. 2011. PMID: 21518905 Free PMC article. - The autophagy effector Beclin 1: a novel BH3-only protein.
Sinha S, Levine B. Sinha S, et al. Oncogene. 2008 Dec;27 Suppl 1(Suppl 1):S137-48. doi: 10.1038/onc.2009.51. Oncogene. 2008. PMID: 19641499 Free PMC article. Review. - Impaired autophagy and APP processing in Alzheimer's disease: The potential role of Beclin 1 interactome.
Salminen A, Kaarniranta K, Kauppinen A, Ojala J, Haapasalo A, Soininen H, Hiltunen M. Salminen A, et al. Prog Neurobiol. 2013 Jul-Aug;106-107:33-54. doi: 10.1016/j.pneurobio.2013.06.002. Epub 2013 Jul 1. Prog Neurobiol. 2013. PMID: 23827971 Review.
Cited by
- BECN1/Beclin 1 sorts cell-surface APP/amyloid β precursor protein for lysosomal degradation.
Swaminathan G, Zhu W, Plowey ED. Swaminathan G, et al. Autophagy. 2016 Dec;12(12):2404-2419. doi: 10.1080/15548627.2016.1234561. Epub 2016 Oct 7. Autophagy. 2016. PMID: 27715386 Free PMC article. - Perturbation of Cellular Redox Homeostasis Dictates Divergent Effects of Polybutyl Cyanoacrylate (PBCA) Nanoparticles on Autophagy.
Sønstevold T, Engedal N, Torgersen ML. Sønstevold T, et al. Cells. 2021 Dec 6;10(12):3432. doi: 10.3390/cells10123432. Cells. 2021. PMID: 34943939 Free PMC article. - CircCBFB is a mediator of hepatocellular carcinoma cell autophagy and proliferation through miR-424-5p/ATG14 axis.
Zhao Z, He J, Feng C. Zhao Z, et al. Immunol Res. 2022 Jun;70(3):341-353. doi: 10.1007/s12026-021-09255-8. Epub 2022 Jan 23. Immunol Res. 2022. PMID: 35066780 - Posttranslational modification of autophagy-related proteins in macroautophagy.
Xie Y, Kang R, Sun X, Zhong M, Huang J, Klionsky DJ, Tang D. Xie Y, et al. Autophagy. 2015;11(1):28-45. doi: 10.4161/15548627.2014.984267. Autophagy. 2015. PMID: 25484070 Free PMC article. Review. - Regulation of Beclin 1 Protein Phosphorylation and Autophagy by Protein Phosphatase 2A (PP2A) and Death-associated Protein Kinase 3 (DAPK3).
Fujiwara N, Usui T, Ohama T, Sato K. Fujiwara N, et al. J Biol Chem. 2016 May 13;291(20):10858-66. doi: 10.1074/jbc.M115.704908. Epub 2016 Mar 18. J Biol Chem. 2016. PMID: 26994142 Free PMC article.
References
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10:458–467 - PubMed
- Komatsu M, Ichimura Y. 2010. Selective autophagy regulates various cellular functions. Genes Cells 15:923–933 - PubMed
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402:672–676 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials