Ubiquitin interacts with the Tollip C2 and CUE domains and inhibits binding of Tollip to phosphoinositides - PubMed (original) (raw)
Ubiquitin interacts with the Tollip C2 and CUE domains and inhibits binding of Tollip to phosphoinositides
Sharmistha Mitra et al. J Biol Chem. 2013.
Abstract
A large number of cellular signaling processes are directed through internalization, via endocytosis, of polyubiquitinated cargo proteins. Tollip is an adaptor protein that facilitates endosomal cargo sorting for lysosomal degradation. Tollip preferentially binds phosphatidylinositol 3-phosphate (PtdIns(3)P) via its C2 domain, an association that may be required for endosomal membrane targeting. Here, we show that Tollip binds ubiquitin through its C2 and CUE domains and that its association with the C2 domain inhibits PtdIns(3)P binding. NMR analysis demonstrates that the C2 and CUE domains bind to overlapping sites on ubiquitin, suggesting that two ubiquitin molecules associate with Tollip simultaneously. Hydrodynamic studies reveal that ubiquitin forms heterodimers with the CUE domain, indicating that the association disrupts the dimeric state of the CUE domain. We propose that, in the absence of polyubiquitinated cargo, the dual binding of ubiquitin partitions Tollip into membrane-bound and membrane-free states, a function that contributes to the engagement of Tollip in both membrane trafficking and cytosolic pathways.
Keywords: Endosomes; Membrane Trafficking; Nuclear Magnetic Resonance; Phosphoinositides; Ubiquitin.
Figures
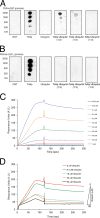
FIGURE 1.
Ubiquitin inhibits the PtdIns(3)P binding of Tollip. Lipid-protein overlay assay of the indicated proteins with immobilized PtdIns(3)P (A) or PtdIns(4,5)P2 (B). To test ubiquitin function, GST-Tollip was preincubated with ubiquitin at the indicated molar ratios for 1 h at room temperature. GST and GST-ubiquitin were employed as negative controls. C, representative SPR sensorgram for the binding of GST-Tollip to PtdIns(3)P-containing liposomes. Various indicated concentrations of GST-Tollip were flown over immobilized PtdIns(3)P liposomes. D, kinetics of ubiquitin-mediated inhibition of Tollip-PtdIns(3)P association. Tollip was preincubated with ubiquitin at the indicated concentrations and association of Tollip to immobilized PtdIns(3)P liposomes was followed using SPR.
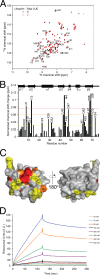
FIGURE 2.
The Tollip C2 domain is an ubiquitin-binding domain. A, lipid-protein overlay assay of the indicated proteins with immobilized PtdIns(3)P. To test ubiquitin function, the Tollip C2 and CUE domains were preincubated with ubiquitin at the indicated molar ratios for 1 h at room temperature. B, lipid-protein overlay assay of the Vam7p PX domain with immobilized PtdIns(3)P and PKCα and PKCβ II C2 domains with immobilized PtdSer in the absence and presence of ubiquitin. C, representative SPR sensorgram for binding of the Tollip C2 domain to immobilized ubiquitin. Various concentrations of Tollip C2 were flown over His-tagged ubiquitin attached on an NTA sensor chip. D, identification of the ubiquitin residues involved in Tollip C2 domain binding. 15N-Labeled ubiquitin was subjected to HSQC analysis in the absence (black) and presence (red) of the Tollip C2 domain. Perturbed ubiquitin resonances are boxed. E, histogram identifying ubiquitin critical residues for Tollip C2 domain recognition. The colored dashed lines represent significant changes, based on the magnitude of their associated chemical shifts changes: red (Δδaverage + 1.5 × S.D.) > orange (Δδaverage + 1 × S.D.) > yellow (Δδaverage). F, two different views of ubiquitin showing the residues involved in Tollip C2 domain binding and color-coded according to the scales defined in E.
FIGURE 3.
Identification of ubiquitin residues involved in Tollip CUE domain binding. A, 15N-labeled ubiquitin was subjected to HSQC analysis in the absence (black) and presence (red) of the Tollip CUE domain. Perturbed ubiquitin resonances are boxed. B, histogram identifying ubiquitin critical residues in Tollip CUE domain recognition. Chemical shift perturbations are classified as indicated in the legend to Fig. 2. C, two different views of ubiquitin showing the residues involved in Tollip CUE domain binding and color-coded according to the scales defined in B. D, representative SPR sensorgram for the binding of GST-Tollip to immobilized His-ubiquitin. Various indicated concentrations of GST-Tollip were flown over immobilized His-ubiquitin.
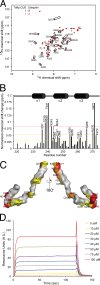
FIGURE 4.
Identification of the Tollip CUE domain residues involved in ubiquitin binding. A, 15N-labeled Tollip CUE domain was subjected to HSQC analysis in the absence (black) and presence (red) of ubiquitin. Perturbed resonances of the Tollip CUE domain are boxed. B, the histogram shows normalized chemical shift perturbations in the backbone amides of the CUE domain induced by ubiquitin. Chemical shift perturbations are classified as indicated in the legend to Fig. 2. C, residues that exhibit significant chemical shift perturbations in A are labeled on the modeled Tollip CUE domain surface and color-coded according to the scales defined in B. D, representative SPR sensorgram for the binding of Tollip CUE domain to immobilized ubiquitin. Various concentrations of the Tollip CUE were flown over His-ubiquitin attached on an nitrilotriacetic acid sensor chip.
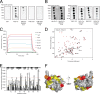
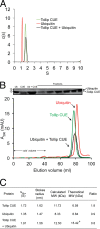
FIGURE 5.
Hydrodynamic properties of the Tollip CUE-ubiquitin complex. A, representative sedimentation velocity analysis of the Tollip CUE-ubiquitin complex. Sedimentation coefficient distribution of free Tollip CUE (_s_app = 1.64; green line), free ubiquitin (_s_app = 1.18; red line), and Tollip CUE-ubiquitin complex (_s_app = 1.73; black line). B, representative gel filtration analysis of ubiquitin (red), Tollip CUE domain (green), and ubiquitin:Tollip CUE domain (1:1 molar ratio; black) using a Superdex 75 column. Fractions of each of the peaks were analyzed using SDS-PAGE (top). C, summary of the results obtained from sedimentation velocity ultracentrifugation and analytical gel filtration analyses.
FIGURE 6.
A proposed model for the regulation of the endosomal membrane-associated Tollip. Tollip cycles between ubiquitin-free and -bound states in the absence of cargo proteins. Endosomal membrane binding of Tollip is mediated by the interaction of its C2 domain with PtdIns(3)P. The CUE domain mediates Tollip dimerization and this event and PtdIns(3)P binding is negatively regulated by ubiquitin. Of note, other ubiquitin-independent regions in Tollip (e.g. TBD) may also contribute to Tollip oligomerization.
Similar articles
- Protein Trafficking or Cell Signaling: A Dilemma for the Adaptor Protein TOM1.
Roach TG, Lång HKM, Xiong W, Ryhänen SJ, Capelluto DGS. Roach TG, et al. Front Cell Dev Biol. 2021 Feb 26;9:643769. doi: 10.3389/fcell.2021.643769. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33718385 Free PMC article. Review. - Tom1 Modulates Binding of Tollip to Phosphatidylinositol 3-Phosphate via a Coupled Folding and Binding Mechanism.
Xiao S, Brannon MK, Zhao X, Fread KI, Ellena JF, Bushweller JH, Finkielstein CV, Armstrong GS, Capelluto DGS. Xiao S, et al. Structure. 2015 Oct 6;23(10):1910-1920. doi: 10.1016/j.str.2015.07.017. Epub 2015 Aug 27. Structure. 2015. PMID: 26320582 - The C2 domain of Tollip, a Toll-like receptor signalling regulator, exhibits broad preference for phosphoinositides.
Ankem G, Mitra S, Sun F, Moreno AC, Chutvirasakul B, Azurmendi HF, Li L, Capelluto DG. Ankem G, et al. Biochem J. 2011 May 1;435(3):597-608. doi: 10.1042/BJ20102160. Biochem J. 2011. PMID: 21294713 - Intracellular trafficking of interleukin-1 receptor I requires Tollip.
Brissoni B, Agostini L, Kropf M, Martinon F, Swoboda V, Lippens S, Everett H, Aebi N, Janssens S, Meylan E, Felberbaum-Corti M, Hirling H, Gruenberg J, Tschopp J, Burns K. Brissoni B, et al. Curr Biol. 2006 Nov 21;16(22):2265-70. doi: 10.1016/j.cub.2006.09.062. Curr Biol. 2006. PMID: 17113392 - Tollip: a multitasking protein in innate immunity and protein trafficking.
Capelluto DG. Capelluto DG. Microbes Infect. 2012 Feb;14(2):140-7. doi: 10.1016/j.micinf.2011.08.018. Epub 2011 Sep 8. Microbes Infect. 2012. PMID: 21930231 Review.
Cited by
- Protein Trafficking or Cell Signaling: A Dilemma for the Adaptor Protein TOM1.
Roach TG, Lång HKM, Xiong W, Ryhänen SJ, Capelluto DGS. Roach TG, et al. Front Cell Dev Biol. 2021 Feb 26;9:643769. doi: 10.3389/fcell.2021.643769. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33718385 Free PMC article. Review. - Toll-Interacting Protein in Resolving and Non-Resolving Inflammation.
Kowalski EJA, Li L. Kowalski EJA, et al. Front Immunol. 2017 May 5;8:511. doi: 10.3389/fimmu.2017.00511. eCollection 2017. Front Immunol. 2017. PMID: 28529512 Free PMC article. Review. - Ubiquitination in the ERAD Process.
Lopata A, Kniss A, Löhr F, Rogov VV, Dötsch V. Lopata A, et al. Int J Mol Sci. 2020 Jul 28;21(15):5369. doi: 10.3390/ijms21155369. Int J Mol Sci. 2020. PMID: 32731622 Free PMC article. Review. - Endocytic Adaptor Protein Tollip Inhibits Canonical Wnt Signaling.
Toruń A, Szymańska E, Castanon I, Wolińska-Nizioł L, Bartosik A, Jastrzębski K, Miętkowska M, González-Gaitán M, Miaczynska M. Toruń A, et al. PLoS One. 2015 Jun 25;10(6):e0130818. doi: 10.1371/journal.pone.0130818. eCollection 2015. PLoS One. 2015. PMID: 26110841 Free PMC article. - Signal Transduction and Intracellular Trafficking by the Interleukin 36 Receptor.
Saha SS, Singh D, Raymond EL, Ganesan R, Caviness G, Grimaldi C, Woska JR Jr, Mennerich D, Brown SE, Mbow ML, Kao CC. Saha SS, et al. J Biol Chem. 2015 Sep 25;290(39):23997-4006. doi: 10.1074/jbc.M115.653378. Epub 2015 Aug 12. J Biol Chem. 2015. PMID: 26269592 Free PMC article.
References
- Husnjak K., Dikic I. (2012) Ubiquitin-binding proteins. Decoders of ubiquitin-mediated cellular functions. Ann. Rev. Biochem. 81, 291–322 - PubMed
- Ikeda F., Crosetto N., Dikic I. (2010) What determines the specificity and outcomes of ubiquitin signaling? Cell 143, 677–681 - PubMed
- Lange O. F., Lakomek N. A., Farès C., Schröder G. F., Walter K. F., Becker S., Meiler J., Grubmüller H., Griesinger C., de Groot B. L. (2008) Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science 320, 1471–1475 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous