Rhesus monkey TRIM5α SPRY domain recognizes multiple epitopes that span several capsid monomers on the surface of the HIV-1 mature viral core - PubMed (original) (raw)
Rhesus monkey TRIM5α SPRY domain recognizes multiple epitopes that span several capsid monomers on the surface of the HIV-1 mature viral core
Nikolaos Biris et al. J Mol Biol. 2013.
Abstract
The restriction factor TRIM5α binds to the capsid protein of the retroviral core and blocks retroviral replication. The affinity of TRIM5α for the capsid is a major host tropism determinant of HIV and other primate immunodeficiency viruses, but the molecular interface involved in this host-pathogen interaction remains poorly characterized. Here we use NMR spectroscopy to investigate binding of the rhesus TRIM5α SPRY domain to a selection of HIV capsid constructs. The data are consistent with a model in which one SPRY domain interacts with more than one capsid monomer within the assembled retroviral core. The highly mobile SPRY v1 loop appears to span the gap between neighboring capsid hexamers making interhexamer contacts critical for restriction. The interaction interface is extensive, involves mobile loops and multiple epitopes, and lacks interaction hot spots. These properties, which may enhance resistance of TRIM5α to capsid mutations, result in relatively low affinity of the individual SPRY domains for the capsid, and the TRIM5α-mediated restriction depends on the avidity effect arising from the oligomerization of TRIM5α.
Keywords: HIV; HSQC; NMR; SPRY; TRIM5α; TROSY; heteronuclear single quantum coherence; transverse relaxation optimized spectroscopy.
© 2013.
Figures
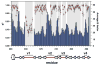
Fig. 1
NMR assignments and protein mobility. Comparison plot of the amide order parameters _S_2 determined using model-free approach (red circles) and the average _B_-factors (blue bars) for the backbone heavy atoms (Cα, C′, and N). Protein segments with missing NMR assignments appear as white gaps in the gray background.
Fig. 2
v1 mobility and the associated primary sequence patterns. (a) Superposition of one of the TRIM5 SPRY structures from the NMR ensemble (green, yellow side chains; PDB code: 2LM3) and the TRIM21 SPRY crystal structure (blue, cyan side chains; PDB code: 2IWG). F325 anchors the v1 segment to the protein core in the TRIM21 structure, whereas in the TRIM5 SPRY, the analogous pocket is occupied by the v2 residue F383. (b) Virtually all of the primate TRIM5 variants have an aromatic or large hydrophobic residue in the position corresponding to F383 of the rhesus TRIM5 (highlighted yellow). In contrast, most of the other SPRY-containing TRIM proteins in the human genome contain a small or polar/charged residue in this position (highlighted orange), which correlates with the occurrence of an RF motif in v1 (highlighted blue). Abbreviations: RH, rhesus monkey, Macaca mulatta; SA, white-lipped tamarin, Saguinus labiatus; CA, white-fronted capuchin, Cebus albifrons; BH, hoolock gibbon, Bunopithecus hoolock; EP, patas monkey, Erythrocebus patas; HU, Homo sapiens.
Fig. 3
Signal attenuation patterns observed in NMR titrations of the 15N-labeled SPRY domain with unlabeled capsid constructs. (a) An area of an 15N TROSY spectrum of the free rhesus TRIM5α SPRY domain with some of the signals corresponding to v1 residues labeled. The same spectrum after addition of the 2:1 molar excess of the CA-NTD (b) or CA-HEX (c) capsid constructs. Enhanced attenuation of the v1 signals is observed in the CA-NTD titration, whereas in the CA-HEX titration, the v1 signals are broadened less than the rest of the protein.
Fig. 4
Sample NMR attenuation curves. Distinct broadening effects of the CA-NTD and CA-HEX constructs on the SPRY spectra are revealed when NMR signal intensities of individual residues are plotted as a function of capsid concentration. In the CA-NTD titration (open circles and triangles), the broadening rate of the residue N345 (triangles) located in the v1 loop is higher than the average protein signal broadening rate (circles). The effect is the opposite in the CA-HEX titration (filled circles and triangles).
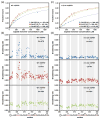
Fig. 5
NMR titrations of the 15N-labeled SPRY. The wild-type rhesus SPRY construct (a and b) and the v1C-5A mutant SPRY (c and d) were titrated with three different capsid constructs: CA-NTD, CA-WT, and CA-HEX. Capsid concentration is given as the concentration of the capsid monomer for all titrations. The intensity attenuation upon addition of increasing amounts of capsid was converted to fraction bound as described in Materials and Methods. (a and c) The binding curves and the derived apparent dissociation constructs determined in the titrations of the WT SPRY (a) and v1C-5A SPRY (c) with the three capsid constructs. (b and d) The broadening rates of the backbone amide NMR signals plotted for all assigned SPRY residues. The enhanced broadening of the signals in the loops v1 and v2 is apparent when the WT SPRY is titrated with the CA-NTD and CA-WT construct, whereas it is not observed in the CA-HEX titration. The enhanced v1/v2 broadening is also not observed in the titrations performed with the v1C-5A SPRY mutant.
Fig. 6
NMR titrations of the perdeuterated, 15N-labeled CA-WT. Titrations were performed and analyzed in exactly the same way as the titrations of the 15N-labeled SPRY. (a) The broadening rate plot reveals that a subset of CA-WT residues displays enhanced signal attenuation. (b) In the crystal structure of the capsid hexamer, residues with elevated broadening rates (red and orange) map onto the outer edge of the hexamer.
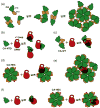
Fig. 7
Low-resolution model of the SPRY–capsid interaction. (a) HIV capsid contains two independently folded domains CA-NTD and CA-CTD connected by a flexible linker. The CA-CTD-mediated dimerization and CA-NTD-mediated hexamerization are two major interfaces formed in the hexagonal-like assembly of the capsid in the mature viral core. (b) All the epitopes recognized by the SPRY domain are located within the CA-NTD. Our data suggest that there are several distinct and possibly overlapping CA-NTD epitopes that the SPRY can possibly bind to. (c) Dimerization of CA-WT can mimic relative orientation of two CA-NTDs located in the neighboring hexamers within the assembled core. Binding of the SPRY domain to CA-WT involves the v1 loop and has higher affinity than binding to the isolated CA-NTD. (d) Isolated capsid hexamers (CA-HEX) produced by disulfide cross-linking of the CA-NTD (yellow bars) and by disruption of the CA-CTD dimerization interface (red stars) binds to the SPRY with higher affinity than CA-NTD, but the binding does not involve the v1 loop. (e) Our data are best explained by a binding model that positions the SPRY domain over the interhexamer gap with the v1 loop spanning the gap. (f) Alternative model to account for the lack of v1 involvement in binding to CA-HEX. v1 may interact with capsid surfaces that are no longer accessible in the cross-linked hexamer.
Similar articles
- Rhesus TRIM5α disrupts the HIV-1 capsid at the inter-hexamer interfaces.
Zhao G, Ke D, Vu T, Ahn J, Shah VB, Yang R, Aiken C, Charlton LM, Gronenborn AM, Zhang P. Zhao G, et al. PLoS Pathog. 2011 Mar;7(3):e1002009. doi: 10.1371/journal.ppat.1002009. Epub 2011 Mar 24. PLoS Pathog. 2011. PMID: 21455494 Free PMC article. - General Model for Retroviral Capsid Pattern Recognition by TRIM5 Proteins.
Wagner JM, Christensen DE, Bhattacharya A, Dawidziak DM, Roganowicz MD, Wan Y, Pumroy RA, Demeler B, Ivanov DN, Ganser-Pornillos BK, Sundquist WI, Pornillos O. Wagner JM, et al. J Virol. 2018 Jan 30;92(4):e01563-17. doi: 10.1128/JVI.01563-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29187540 Free PMC article. - Recognition of the HIV capsid by the TRIM5α restriction factor is mediated by a subset of pre-existing conformations of the TRIM5α SPRY domain.
Kovalskyy DB, Ivanov DN. Kovalskyy DB, et al. Biochemistry. 2014 Mar 11;53(9):1466-76. doi: 10.1021/bi4014962. Epub 2014 Feb 24. Biochemistry. 2014. PMID: 24506064 Free PMC article. - Impact of TRIM5α in vivo.
Nakayama EE, Shioda T. Nakayama EE, et al. AIDS. 2015 Sep 10;29(14):1733-43. doi: 10.1097/QAD.0000000000000812. AIDS. 2015. PMID: 26372380 Free PMC article. Review. - Anti-retroviral activity of TRIM5 alpha.
Nakayama EE, Shioda T. Nakayama EE, et al. Rev Med Virol. 2010 Mar;20(2):77-92. doi: 10.1002/rmv.637. Rev Med Virol. 2010. PMID: 20049904 Review.
Cited by
- Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice.
Goldstone DC, Walker PA, Calder LJ, Coombs PJ, Kirkpatrick J, Ball NJ, Hilditch L, Yap MW, Rosenthal PB, Stoye JP, Taylor IA. Goldstone DC, et al. Proc Natl Acad Sci U S A. 2014 Jul 1;111(26):9609-14. doi: 10.1073/pnas.1402448111. Epub 2014 Jun 16. Proc Natl Acad Sci U S A. 2014. PMID: 24979782 Free PMC article. - Ring finger protein 39 genetic variants associate with HIV-1 plasma viral loads and its replication in cell culture.
Lin YJ, Chen CY, Jeang KT, Liu X, Wang JH, Hung CH, Tsang H, Lin TH, Liao CC, Huang SM, Lin CW, Ho MW, Chien WK, Chen JH, Ho TJ, Tsai FJ. Lin YJ, et al. Cell Biosci. 2014 Aug 5;4:40. doi: 10.1186/2045-3701-4-40. eCollection 2014. Cell Biosci. 2014. PMID: 25126410 Free PMC article. - Antiviral innate immunity: editorial overview.
Freed EO, Gale M Jr. Freed EO, et al. J Mol Biol. 2014 Mar 20;426(6):1129-32. doi: 10.1016/j.jmb.2014.01.005. Epub 2014 Jan 22. J Mol Biol. 2014. PMID: 24462565 Free PMC article. No abstract available. - Restriction of HIV-1 and other retroviruses by TRIM5.
Ganser-Pornillos BK, Pornillos O. Ganser-Pornillos BK, et al. Nat Rev Microbiol. 2019 Sep;17(9):546-556. doi: 10.1038/s41579-019-0225-2. Epub 2019 Jul 16. Nat Rev Microbiol. 2019. PMID: 31312031 Free PMC article. Review. - The small-molecule 3G11 inhibits HIV-1 reverse transcription.
Opp S, Fricke T, Shepard C, Kovalskyy D, Bhattacharya A, Herkules F, Ivanov DN, Kim B, Valle-Casuso J, Diaz-Griffero F. Opp S, et al. Chem Biol Drug Des. 2017 Apr;89(4):608-618. doi: 10.1111/cbdd.12886. Epub 2016 Nov 15. Chem Biol Drug Des. 2017. PMID: 27748043 Free PMC article.
References
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–53. - PubMed
- Ganser-Pornillos BK, Cheng A, Yeager M. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell. 2007;131:70–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 GM082545/GM/NIGMS NIH HHS/United States
- R21 AI084612/AI/NIAID NIH HHS/United States
- CA054174/CA/NCI NIH HHS/United States
- R01 AI087390/AI/NIAID NIH HHS/United States
- R01 AI104476/AI/NIAID NIH HHS/United States
- P30 CA054174/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous