A microfluidic DNA library preparation platform for next-generation sequencing - PubMed (original) (raw)
. 2013 Jul 22;8(7):e68988.
doi: 10.1371/journal.pone.0068988. Print 2013.
Mais J Jebrail, Anupama Sinha, Zachary W Bent, Owen D Solberg, Kelly P Williams, Stanley A Langevin, Ronald F Renzi, James L Van De Vreugde, Robert J Meagher, Joseph S Schoeniger, Todd W Lane, Steven S Branda, Michael S Bartsch, Kamlesh D Patel
Affiliations
- PMID: 23894387
- PMCID: PMC3718812
- DOI: 10.1371/journal.pone.0068988
A microfluidic DNA library preparation platform for next-generation sequencing
Hanyoup Kim et al. PLoS One. 2013.
Abstract
Next-generation sequencing (NGS) is emerging as a powerful tool for elucidating genetic information for a wide range of applications. Unfortunately, the surging popularity of NGS has not yet been accompanied by an improvement in automated techniques for preparing formatted sequencing libraries. To address this challenge, we have developed a prototype microfluidic system for preparing sequencer-ready DNA libraries for analysis by Illumina sequencing. Our system combines droplet-based digital microfluidic (DMF) sample handling with peripheral modules to create a fully-integrated, sample-in library-out platform. In this report, we use our automated system to prepare NGS libraries from samples of human and bacterial genomic DNA. E. coli libraries prepared on-device from 5 ng of total DNA yielded excellent sequence coverage over the entire bacterial genome, with >99% alignment to the reference genome, even genome coverage, and good quality scores. Furthermore, we produced a de novo assembly on a previously unsequenced multi-drug resistant Klebsiella pneumoniae strain BAA-2146 (KpnNDM). The new method described here is fast, robust, scalable, and automated. Our device for library preparation will assist in the integration of NGS technology into a wide variety of laboratories, including small research laboratories and clinical laboratories.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
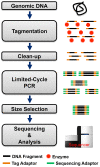
Figure 1. Nextera® protocol for preparing DNA libraries compatible with Illumina sequencers.
Key steps include: gDNA tagmentation, clean-up, limited-cycle PCR, and selection of size-specific DNA library for sequencing and analysis.
Figure 2. Integrated microfluidic system for preparing DNA libraries for sequencing.
a) Top view of an integrated system and side view of the DMF-capillary interface, which features a central DMF hub for integrating multiple reagent and sample preparation modules (depicted in different colors), magnets and thermal blocks coupled to module tubing for sample preparation, and multi-valve syringe pumps for liquid handling. b) Schematic (left) of a DMF device showing the arrangement of indium tin oxide (ITO) actuation electrodes used to route samples and reagents to and from the capillaries, which are responsible for transferring liquids to their respective modules. Sequence of frames (right) from a movie illustratfing the stages in preparing a sequencer-ready DNA library using the microfluidic method (note that off-DMF processing is not shown in the perspective of the frames): (1) Mixing of gDNA (1.8 µL) and Nextera Enzyme (NE, 0.6 µL) droplets; (2,3) post-tagmentation droplet (gDNA+NE reaction products) merged with magnetic bead (MB) droplet (4.5 µL) and actuated to clean-up module; (4,5) post-clean-up droplet (2.2 µL) of purified DNA fragments actuated to PCR Mix droplet (2.8 µL) for limited-cycle PCR; (6,7) post-PCR DNA fragment droplet mixed with different volumes (2.2 and 0.45 µL) of magnetic beads for size-selection; and (8) DNA library droplet (3 µL).
Figure 3. Modular format for implementing magnetic bead-based steps.
Series of images from a movie (side-view) depicting key magnetic bead-based steps in an off-DMF module. Blue- and red-dashed lines show the walls of module and air/bolus interface, respectively. (1, 2) An external magnet immobilizes beads onto the tube surface. (3) Positive pressure-driven flow separates supernatant away from the bead pellet. (4, 5) Removing the magnet and shuttling a bolus of solution over the pellet resuspends immobilized beads. Arrows indicate the direction of bolus movement.
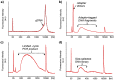
Figure 4. Analysis of human gDNA at different stages of the Nextera protocol.
Bioanalyzer traces of a) gDNA, b) post-tagmentation, c) post-limited-cycle PCR and d) post-size-selection. Peaks at 35 and 10380 bp represent low- and high-molecular weight markers.
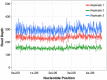
Figure 5. Genome coverage plots for three E. coli libraries prepared by microfluidic method.
The plot represents relative depth of sequence coverage across the entire ∼4.6 kb E. coli genome.
Similar articles
- Automated digital microfluidic sample preparation for next-generation DNA sequencing.
Kim H, Bartsch MS, Renzi RF, He J, Van de Vreugde JL, Claudnic MR, Patel KD. Kim H, et al. J Lab Autom. 2011 Dec;16(6):405-14. doi: 10.1016/j.jala.2011.07.001. Epub 2011 Sep 23. J Lab Autom. 2011. PMID: 22093297 - Microfluidic Platform for Next-Generation Sequencing Library Preparation with Low-Input Samples.
Murphy TW, Hsieh YP, Zhu B, Naler LB, Lu C. Murphy TW, et al. Anal Chem. 2020 Feb 4;92(3):2519-2526. doi: 10.1021/acs.analchem.9b04086. Epub 2020 Jan 14. Anal Chem. 2020. PMID: 31894965 Free PMC article. - Quality control of next-generation sequencing library through an integrative digital microfluidic platform.
Thaitrong N, Kim H, Renzi RF, Bartsch MS, Meagher RJ, Patel KD. Thaitrong N, et al. Electrophoresis. 2012 Dec;33(23):3506-13. doi: 10.1002/elps.201200441. Epub 2012 Nov 8. Electrophoresis. 2012. PMID: 23135807 - Library construction for next-generation sequencing: overviews and challenges.
Head SR, Komori HK, LaMere SA, Whisenant T, Van Nieuwerburgh F, Salomon DR, Ordoukhanian P. Head SR, et al. Biotechniques. 2014 Feb 1;56(2):61-4, 66, 68, passim. doi: 10.2144/000114133. eCollection 2014. Biotechniques. 2014. PMID: 24502796 Free PMC article. Review. - Next-generation sequencing (NGS) in the microbiological world: How to make the most of your money.
Vincent AT, Derome N, Boyle B, Culley AI, Charette SJ. Vincent AT, et al. J Microbiol Methods. 2017 Jul;138:60-71. doi: 10.1016/j.mimet.2016.02.016. Epub 2016 Mar 16. J Microbiol Methods. 2017. PMID: 26995332 Review.
Cited by
- The rotary zone thermal cycler: a low-power system enabling automated rapid PCR.
Bartsch MS, Edwards HS, Lee D, Moseley CE, Tew KE, Renzi RF, Van de Vreugde JL, Kim H, Knight DL, Sinha A, Branda SS, Patel KD. Bartsch MS, et al. PLoS One. 2015 Mar 31;10(3):e0118182. doi: 10.1371/journal.pone.0118182. eCollection 2015. PLoS One. 2015. PMID: 25826708 Free PMC article. - A parallelized microfluidic DNA bisulfite conversion module for streamlined methylation analysis.
Stark A, Shin DJ, Pisanic T 2nd, Hsieh K, Wang TH. Stark A, et al. Biomed Microdevices. 2016 Feb;18(1):5. doi: 10.1007/s10544-015-0029-8. Biomed Microdevices. 2016. PMID: 26759004 Free PMC article. - Microfluidics for genome-wide studies involving next generation sequencing.
Ma S, Murphy TW, Lu C. Ma S, et al. Biomicrofluidics. 2017 Mar 10;11(2):021501. doi: 10.1063/1.4978426. eCollection 2017 Mar. Biomicrofluidics. 2017. PMID: 28396707 Free PMC article. Review. - Microfluidic Technologies for cfDNA Isolation and Analysis.
Xu Z, Qiao Y, Tu J. Xu Z, et al. Micromachines (Basel). 2019 Oct 3;10(10):672. doi: 10.3390/mi10100672. Micromachines (Basel). 2019. PMID: 31623361 Free PMC article. Review. - Integrated magneto-electrophoresis microfluidic chip purification on library preparation device for preimplantation genetic testing for aneuploidy detection.
Schneider L, Fraser M, Tripathi A. Schneider L, et al. RSC Adv. 2021 Apr 16;11(24):14459-14474. doi: 10.1039/d1ra01732b. eCollection 2021 Apr 15. RSC Adv. 2021. PMID: 35423999 Free PMC article.
References
- Metzker ML (2010) Sequencing technologies the next generation. Nat Rev Genet 11: 31–46. - PubMed
- Scholz MB, Lo CC, Chain PSG (2012) Next generation sequencing and bioinformatic bottlenecks: The current state of metagenomic data analysis. Curr Opin Biotechnol 23: 9–15. - PubMed
- Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, et al. (2011) Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 478: 57–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This work was funded by Sandia National Laboratories’ Grand Challenge LDRD (Laboratory-Directed Research and Development, grant number 142042) program. Sandia is a multiprogram laboratory operated by Sandia Corporation, a Lockheed Martin company, for the U.S. Department of Energy’s National Nuclear Security Administration under Contract DE-AC04-94AL85000. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources