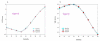MSClust: A Multi-Seeds based Clustering algorithm for microbiome profiling using 16S rRNA sequence - PubMed (original) (raw)
MSClust: A Multi-Seeds based Clustering algorithm for microbiome profiling using 16S rRNA sequence
Wei Chen et al. J Microbiol Methods. 2013 Sep.
Abstract
Recent developments of next generation sequencing technologies have led to rapid accumulation of 16S rRNA sequences for microbiome profiling. One key step in data processing is to cluster short sequences into operational taxonomic units (OTUs). Although many methods have been proposed for OTU inferences, a major challenge is the balance between inference accuracy and computational efficiency, where inference accuracy is often sacrificed to accommodate the need to analyze large numbers of sequences. Inspired by the hierarchical clustering method and a modified greedy network clustering algorithm, we propose a novel multi-seeds based heuristic clustering method, named MSClust, for OTU inference. MSClust first adaptively selects multi-seeds instead of one seed for each candidate cluster, and the reads are then processed using a greedy clustering strategy. Through many numerical examples, we demonstrate that MSClust enjoys less memory usage, and better biological accuracy compared to existing heuristic clustering methods while preserving efficiency and scalability.
Keywords: 16S rRNA reads; Clustering algorithms; Next-generation sequencing; Operational taxonomic unit (OTU); Seeds-selection.
© 2013 Elsevier B.V. All rights reserved.
Figures
Figure 1
Flow chart of the MSClust algorithm
Figure 2
The pseudo-code for the Seeds Selection procedure.
Figure 3
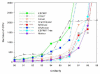
Comparison of the Number of OTUs predicted by ESPRIT, CDHIT, Uclust, GramCluster, MSClust, DNA Clust and ESPRIT-Tree using the V2 benchmark at different sequences similarity thereshould ranging from 0.98 to 0.9
Figure 4
NMI and NID scores for different algorithoms based on the species ground-truth (a). NID scores across different algorithms at different threshoulds. (b). NMI scores across different algorithms at different threshoulds.
Figure 5
The effect of Parameter NumS (Number of Seeds) on clustering results. a). NID scores of MSClust are calculated at various similarity levels with different NumS. b) NMI scores of MSClust are compared for various NumS.
Figure 6
The effect of Parameter Tsize (size of Temp Cluster) on clustering results. a) NID scores of MSClust are calculated at various similarity levels with different Tsize. b) NMI scores of MSClust for various NumS with the same NumS=3.
Figure 7
phylogenetic tree for the 30 species
Figure 8
NID score and NMI score for the simulated data set. (a). NID scores for the compared algorithms at different threshoulds. (b). NMI scores for the compared algorithms at different thresholds.
Figure 9
NMI and NID Scores for different algorithms based on the benchmark dataset Stacked_60 with species ground-truth. (a) NID Scores across different algorithms at different threshoulds. (b) NMI Scores across different algorithms different threshoulds.
Figure 10
Similar articles
- Improved OTU-picking using long-read 16S rRNA gene amplicon sequencing and generic hierarchical clustering.
Franzén O, Hu J, Bao X, Itzkowitz SH, Peter I, Bashir A. Franzén O, et al. Microbiome. 2015 Oct 5;3:43. doi: 10.1186/s40168-015-0105-6. Microbiome. 2015. PMID: 26434730 Free PMC article. - DBH: A de Bruijn graph-based heuristic method for clustering large-scale 16S rRNA sequences into OTUs.
Wei ZG, Zhang SW. Wei ZG, et al. J Theor Biol. 2017 Jul 21;425:80-87. doi: 10.1016/j.jtbi.2017.04.019. Epub 2017 Apr 26. J Theor Biol. 2017. PMID: 28454900 - MtHc: a motif-based hierarchical method for clustering massive 16S rRNA sequences into OTUs.
Wei ZG, Zhang SW. Wei ZG, et al. Mol Biosyst. 2015 Jul;11(7):1907-13. doi: 10.1039/c5mb00089k. Mol Biosyst. 2015. PMID: 25912934 - bioOTU: An Improved Method for Simultaneous Taxonomic Assignments and Operational Taxonomic Units Clustering of 16s rRNA Gene Sequences.
Chen SY, Deng F, Huang Y, Jia X, Liu YP, Lai SJ. Chen SY, et al. J Comput Biol. 2016 Apr;23(4):229-38. doi: 10.1089/cmb.2015.0214. Epub 2016 Mar 7. J Comput Biol. 2016. PMID: 26950196 - DMclust, a Density-based Modularity Method for Accurate OTU Picking of 16S rRNA Sequences.
Wei ZG, Zhang SW, Zhang YZ. Wei ZG, et al. Mol Inform. 2017 Dec;36(12). doi: 10.1002/minf.201600059. Epub 2017 Jun 6. Mol Inform. 2017. PMID: 28586119
Cited by
- Toward accurate molecular identification of species in complex environmental samples: testing the performance of sequence filtering and clustering methods.
Flynn JM, Brown EA, Chain FJ, MacIsaac HJ, Cristescu ME. Flynn JM, et al. Ecol Evol. 2015 Jun;5(11):2252-66. doi: 10.1002/ece3.1497. Epub 2015 May 13. Ecol Evol. 2015. PMID: 26078860 Free PMC article. - ESPRIT-Forest: Parallel clustering of massive amplicon sequence data in subquadratic time.
Cai Y, Zheng W, Yao J, Yang Y, Mai V, Mao Q, Sun Y. Cai Y, et al. PLoS Comput Biol. 2017 Apr 24;13(4):e1005518. doi: 10.1371/journal.pcbi.1005518. eCollection 2017 Apr. PLoS Comput Biol. 2017. PMID: 28437450 Free PMC article. - A parallel computational framework for ultra-large-scale sequence clustering analysis.
Zheng W, Mao Q, Genco RJ, Wactawski-Wende J, Buck M, Cai Y, Sun Y. Zheng W, et al. Bioinformatics. 2019 Feb 1;35(3):380-388. doi: 10.1093/bioinformatics/bty617. Bioinformatics. 2019. PMID: 30010718 Free PMC article. - Improved OTU-picking using long-read 16S rRNA gene amplicon sequencing and generic hierarchical clustering.
Franzén O, Hu J, Bao X, Itzkowitz SH, Peter I, Bashir A. Franzén O, et al. Microbiome. 2015 Oct 5;3:43. doi: 10.1186/s40168-015-0105-6. Microbiome. 2015. PMID: 26434730 Free PMC article. - DMSC: A Dynamic Multi-Seeds Method for Clustering 16S rRNA Sequences Into OTUs.
Wei ZG, Zhang SW. Wei ZG, et al. Front Microbiol. 2019 Mar 12;10:428. doi: 10.3389/fmicb.2019.00428. eCollection 2019. Front Microbiol. 2019. PMID: 30915052 Free PMC article.
References
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbial. 2009;75(23):7537–7541. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources