HIV-1 capsid undergoes coupled binding and isomerization by the nuclear pore protein NUP358 - PubMed (original) (raw)
HIV-1 capsid undergoes coupled binding and isomerization by the nuclear pore protein NUP358
Katsiaryna Bichel et al. Retrovirology. 2013.
Abstract
Background: Lentiviruses such as HIV-1 can be distinguished from other retroviruses by the cyclophilin A-binding loop in their capsid and their ability to infect non-dividing cells. Infection of non-dividing cells requires transport through the nuclear pore but how this is mediated is unknown.
Results: Here we present the crystal structure of the N-terminal capsid domain of HIV-1 in complex with the cyclophilin domain of nuclear pore protein NUP358. The structure reveals that HIV-1 is positioned to allow single-bond resonance stabilisation of exposed capsid residue P90. NMR exchange experiments demonstrate that NUP358 is an active isomerase, which efficiently catalyzes cis-trans isomerization of the HIV-1 capsid. In contrast, the distantly related feline lentivirus FIV can bind NUP358 but is neither isomerized by it nor requires it for infection.
Conclusion: Isomerization by NUP358 may be preserved by HIV-1 to target the nuclear pore and synchronize nuclear entry with capsid uncoating.
Figures
Figure 1
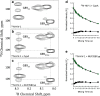
HIV-1 CA N :NUP358Cyp complex. (a) Sequence alignment of NUP358Cyp with CypA (conserved regions are shaded and in bold). (b) Structural alignment of NUP358Cyp (yellow) from our structure with CypA (gray) from the HIV-1 CAN:CypA structure (pdb 1AK4 [23]. (c) HIV-1 CAN (green) bound to CypA (molecular surface; gray) (pdb 1AK4 [23]). (d) HIV-1 CAN (green) bound to NUP358Cyp (molecular surface; yellow). (e) Detailed view of interactions between the CypA-binding loop of HIV-1 CAN (green ball-and-stick) and CypA (gray). (f) Detailed view of interactions between the CypA-binding loop of HIV-1 CAN (green ball-and-stick) and NUP358Cyp (yellow). NUP358Cyp residues are numbered according to the equivalent CypA numbering. (g-h) Structure of CypA:Cs complex (2RMA [24]) (g) or model where NUP358Cyp has been substituted for CypA (h). Cs is in green.
Figure 2
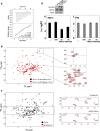
Co-operative NUP358 residues determine HIV-1 capsid binding. (a) Sequence alignment of NUP358Cyp from primates and other mammals. Residues at position 61 and 113 are highlighted. (b) ITC traces for HIV-1 CAN binding to NUP358Cyp mutants. The location of mutated residues are indicated in the figures below each trace, with residues 61 and 113 shown as space-filling spheres. CypA is indicated in gray and NUP358Cyp in yellow. (c) Structural alignment of CypA (gray) from HIV-1 CAN:CypA complex 1AK4 with HIV-1 CAN:NUP358Cyp (yellow). The CypA-binding loop from HIV-1 CAN is shown in green. Divergent residues in proximity to the binding site are indicated.
Figure 3
NUP358Cyp catalyses cis-trans isomerization of HIV-1 CA N . (a-c) 2D 1H-15N ZZ-exchange spectra of HIV-1 CAN focused on Gly89, without enzyme (a), in the presence of NUP358Cyp (b) or CypA (c). Gly89 _cis,trans_1H,15N correlation or “auto” peaks are labeled and are the result of cis and trans forms of the proceeding Pro90 residue being populated at equilibrium. Addition of either NUP358Cyp or CypA yields exchange peaks that connect cis and trans auto peaks (broken lines). The cis exchange peak appears at the same 1H (15N) chemical shift position as the trans auto peak and vice versa. Note, that the trans exchange peak is largely obscured by additional signals in the spectra. (d-e) The intensities of both auto and exchange peaks vary as a result of the ZZ-mixing time (Tm). Fits of CypA (d) and NUP358Cyp (e) yield exchange constants of 4.3 and 12.1 s-1 respectively.
Figure 4
FIV uncouples binding and isomerization by NUP358. (a) ITC trace of NUP358Cyp interaction with FIV CAN. (b) Overlay of 2D 1H-15N ZZ-exchange spectra of FIV CAN in the presence of NUP358Cyp at 3 and 197 ms mixing times. Zoomed-in views of the peaks corresponding to G87 and R89 are also shown. The cross-hairs indicate where exchange peaks would be situated, if cis-trans isomerization took place. (c) Overlay of uniformly 15N-labeled and selective 15N-Arg labeled FIV CAN. Spectra of selective 15N-Arg labeled FIV CAN are also shown in panels on the right in the absence (top) and presence (bottom) of human NUP358Cyp. (d) Upper: Western blots showing depletion of NUP358 or TNPO3 in HeLa cells using antibodies against NUP358, TNPO3 or β-Actin as loading control [6]. Lower: VSV-G pseudotyped GFP-encoding HIV-1 or FIV infection of HeLa cells and HeLa cells expressing scrambled control (shC) or NUP358 or TNPO3 specific shRNA [6,33] (mean of three independent viral doses, error bars show standard deviation). Titers are plotted as infectious units per ng of reverse transcriptase activity. Data are representative of two independent experiments.
Similar articles
- The Role of Capsid in HIV-1 Nuclear Entry.
Guedán A, Caroe ER, Barr GCR, Bishop KN. Guedán A, et al. Viruses. 2021 Jul 22;13(8):1425. doi: 10.3390/v13081425. Viruses. 2021. PMID: 34452291 Free PMC article. Review. - Structural and functional analysis of the C-terminal domain of Nup358/RanBP2.
Lin DH, Zimmermann S, Stuwe T, Stuwe E, Hoelz A. Lin DH, et al. J Mol Biol. 2013 Apr 26;425(8):1318-29. doi: 10.1016/j.jmb.2013.01.021. Epub 2013 Jan 23. J Mol Biol. 2013. PMID: 23353830 Free PMC article. - HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency.
Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, Hué S, Fletcher AJ, Lee K, KewalRamani VN, Noursadeghi M, Jenner RG, James LC, Bushman FD, Towers GJ. Schaller T, et al. PLoS Pathog. 2011 Dec;7(12):e1002439. doi: 10.1371/journal.ppat.1002439. Epub 2011 Dec 8. PLoS Pathog. 2011. PMID: 22174692 Free PMC article. - KIF5B and Nup358 Cooperatively Mediate the Nuclear Import of HIV-1 during Infection.
Dharan A, Talley S, Tripathi A, Mamede JI, Majetschak M, Hope TJ, Campbell EM. Dharan A, et al. PLoS Pathog. 2016 Jun 21;12(6):e1005700. doi: 10.1371/journal.ppat.1005700. eCollection 2016 Jun. PLoS Pathog. 2016. PMID: 27327622 Free PMC article. - A model for cofactor use during HIV-1 reverse transcription and nuclear entry.
Hilditch L, Towers GJ. Hilditch L, et al. Curr Opin Virol. 2014 Feb;4(100):32-6. doi: 10.1016/j.coviro.2013.11.003. Epub 2014 Jan 14. Curr Opin Virol. 2014. PMID: 24525292 Free PMC article. Review.
Cited by
- HIV-1 adapts to lost IP6 coordination through second-site mutations that restore conical capsid assembly.
Kleinpeter A, Mallery DL, Renner N, Albecka A, Klarhof JO, Freed EO, James LC. Kleinpeter A, et al. Nat Commun. 2024 Sep 13;15(1):8017. doi: 10.1038/s41467-024-51971-w. Nat Commun. 2024. PMID: 39271696 Free PMC article. - Tough Way In, Tough Way Out: The Complex Interplay of Host and Viral Factors in Nucleocytoplasmic Trafficking during HIV-1 Infection.
Sarkar S, Balakrishnan K, Chintala K, Mohareer K, Luedde T, Vasudevan AAJ, Münk C, Banerjee S. Sarkar S, et al. Viruses. 2022 Nov 12;14(11):2503. doi: 10.3390/v14112503. Viruses. 2022. PMID: 36423112 Free PMC article. Review. - HIV-1 capsid is involved in post-nuclear entry steps.
Chen NY, Zhou L, Gane PJ, Opp S, Ball NJ, Nicastro G, Zufferey M, Buffone C, Luban J, Selwood D, Diaz-Griffero F, Taylor I, Fassati A. Chen NY, et al. Retrovirology. 2016 Apr 23;13:28. doi: 10.1186/s12977-016-0262-0. Retrovirology. 2016. PMID: 27107820 Free PMC article. - CA Mutation N57A Has Distinct Strain-Specific HIV-1 Capsid Uncoating and Infectivity Phenotypes.
Fischer DK, Saito A, Kline C, Cohen R, Watkins SC, Yamashita M, Ambrose Z. Fischer DK, et al. J Virol. 2019 Apr 17;93(9):e00214-19. doi: 10.1128/JVI.00214-19. Print 2019 May 1. J Virol. 2019. PMID: 30814280 Free PMC article. - The Role of Capsid in HIV-1 Nuclear Entry.
Guedán A, Caroe ER, Barr GCR, Bishop KN. Guedán A, et al. Viruses. 2021 Jul 22;13(8):1425. doi: 10.3390/v13081425. Viruses. 2021. PMID: 34452291 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- G9721629/MRC_/Medical Research Council/United Kingdom
- G0501446/MRC_/Medical Research Council/United Kingdom
- G0900950/MRC_/Medical Research Council/United Kingdom
- G0801172/MRC_/Medical Research Council/United Kingdom
- MC_U105181010/MRC_/Medical Research Council/United Kingdom
- 090940/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous