Functional profiling of the gut microbiome in disease-associated inflammation - PubMed (original) (raw)
Review
Functional profiling of the gut microbiome in disease-associated inflammation
Daniela Börnigen et al. Genome Med. 2013.
Abstract
The microbial residents of the human gut are a major factor in the development and lifelong maintenance of health. The gut microbiota differs to a large degree from person to person and has an important influence on health and disease due to its interaction with the human immune system. Its overall composition and microbial ecology have been implicated in many autoimmune diseases, and it represents a particularly important area for translational research as a new target for diagnostics and therapeutics in complex inflammatory conditions. Determining the biomolecular mechanisms by which altered microbial communities contribute to human disease will be an important outcome of current functional studies of the human microbiome. In this review, we discuss functional profiling of the human microbiome using metagenomic and metatranscriptomic approaches, focusing on the implications for inflammatory conditions such as inflammatory bowel disease and rheumatoid arthritis. Common themes in gut microbial ecology have emerged among these diverse diseases, but they have not yet been linked to targetable mechanisms such as microbial gene and genome composition, pathway and transcript activity, and metabolism. Combining these microbial activities with host gene, transcript and metabolic information will be necessary to understand how and why these complex interacting systems are altered in disease-associated inflammation.
Figures
Figure 1
A model of functional dysbiosis in the human gut microbiome during initiation and progression of complex disease. Although many current studies focus on microbial composition shifts that occur subsequent to disease establishment, it is critical to differentiate functional from structural changes in the microbiome and their distinct patterns in early versus late disease. (a) An illustration of microbial community structural changes during complex disease progression. Ordinations such as principle coordinate analysis and multidimensional scaling are commonly used to qualitatively visualize microbial community structure among multiple samples (for example, cases and controls). Ordinations project distance measures such as beta diversity among samples into fewer dimensions in such a way that the patterns of greatest change occur on the primary axes (here, x and y). However, particularly in early disease, case/control status is frequently not among the factors with most influence on inter-subject microbial variation. Conversely, later-stage inflammation can have a very large effect on microbial structure, causing other sources of variation to become visually less apparent. (b) Functional profiles of gut microbial communities remain more stable among individuals in health than do microbial profiles, and they can likewise show more concerted differential responses in early and late disease stages. In this illustration, 'case' subject samples exhibit expansion of specific metagenomically encoded functions in their microbial communities during progressive phases of inflammation, as reported in [32]. (c) Representative host histology in different phases of the inflammatory response in Crohn's colitis. Colonic crypts (ring structures) are gradually destroyed by immune infiltration as colitis progresses. Images show transverse sections of human colonic mucosa stained with hematoxylin and eosin; 100 µm scale bars are included for reference (images provided by WSG). CDAC, _Clostridium difficile_-associated diarrhea; PC, principal coordinate.
Figure 2
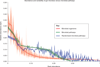
The core gut microbiome consists of stable pathways present despite variation in microbial abundances. Profiles of 118 stool samples from healthy individuals, showing the relative abundances of microbial organisms (red), inferred microbial pathways [70] (green), and microbial pathways after randomization (blue, all data from [1]). All relative abundances are shown as median and interquartile range across all samples (_y_-axis) ranked by median (_x_-axis) and square-root (sqrt) scaled for visualization. As illustrated by several studies (for example, [1,89]), a stable distribution of habitat-adapted microbial pathways is maintained on a functional level (green) rather than on a phylogenetic level (red). Random assignment of microbes to samples followed by re-inference of functional potential (blue) results in a metagenome that is more variable, more skewed, and of distinct composition from that in the observed 'core' of gut microbiome functions.
Similar articles
- Functional dynamics of bacterial species in the mouse gut microbiome revealed by metagenomic and metatranscriptomic analyses.
Chung YW, Gwak HJ, Moon S, Rho M, Ryu JH. Chung YW, et al. PLoS One. 2020 Jan 24;15(1):e0227886. doi: 10.1371/journal.pone.0227886. eCollection 2020. PLoS One. 2020. PMID: 31978162 Free PMC article. - Metabolic phenotyping for understanding the gut microbiome and host metabolic interplay.
Basson AR, Wijeyesekera A. Basson AR, et al. Emerg Top Life Sci. 2017 Nov 30;1(4):325-332. doi: 10.1042/ETLS20170079. Emerg Top Life Sci. 2017. PMID: 33525773 - Metatranscriptomic analysis to define the Secrebiome, and 16S rRNA profiling of the gut microbiome in obesity and metabolic syndrome of Mexican children.
Gallardo-Becerra L, Cornejo-Granados F, García-López R, Valdez-Lara A, Bikel S, Canizales-Quinteros S, López-Contreras BE, Mendoza-Vargas A, Nielsen H, Ochoa-Leyva A. Gallardo-Becerra L, et al. Microb Cell Fact. 2020 Mar 6;19(1):61. doi: 10.1186/s12934-020-01319-y. Microb Cell Fact. 2020. PMID: 32143621 Free PMC article. - The Microbiome in Patients With Inflammatory Diseases.
Somineni HK, Kugathasan S. Somineni HK, et al. Clin Gastroenterol Hepatol. 2019 Jan;17(2):243-255. doi: 10.1016/j.cgh.2018.08.078. Epub 2018 Sep 7. Clin Gastroenterol Hepatol. 2019. PMID: 30196163 Review. - Foodomics as part of the host-microbiota-exposome interplay.
Putignani L, Dallapiccola B. Putignani L, et al. J Proteomics. 2016 Sep 16;147:3-20. doi: 10.1016/j.jprot.2016.04.033. Epub 2016 Apr 26. J Proteomics. 2016. PMID: 27130534 Review.
Cited by
- The Role of High-Mobility Group Box-1 and Its Crosstalk with Microbiome in Rheumatoid Arthritis.
Biscetti F, Flex A, Alivernini S, Tolusso B, Gremese E, Ferraccioli G. Biscetti F, et al. Mediators Inflamm. 2017;2017:5230374. doi: 10.1155/2017/5230374. Epub 2017 Oct 23. Mediators Inflamm. 2017. PMID: 29200665 Free PMC article. Review. - The complexities of the diet-microbiome relationship: advances and perspectives.
Leeming ER, Louca P, Gibson R, Menni C, Spector TD, Le Roy CI. Leeming ER, et al. Genome Med. 2021 Jan 20;13(1):10. doi: 10.1186/s13073-020-00813-7. Genome Med. 2021. PMID: 33472701 Free PMC article. Review. - Functional and phylogenetic assembly of microbial communities in the human microbiome.
Shafquat A, Joice R, Simmons SL, Huttenhower C. Shafquat A, et al. Trends Microbiol. 2014 May;22(5):261-6. doi: 10.1016/j.tim.2014.01.011. Epub 2014 Mar 5. Trends Microbiol. 2014. PMID: 24618403 Free PMC article. Review. - Relating the metatranscriptome and metagenome of the human gut.
Franzosa EA, Morgan XC, Segata N, Waldron L, Reyes J, Earl AM, Giannoukos G, Boylan MR, Ciulla D, Gevers D, Izard J, Garrett WS, Chan AT, Huttenhower C. Franzosa EA, et al. Proc Natl Acad Sci U S A. 2014 Jun 3;111(22):E2329-38. doi: 10.1073/pnas.1319284111. Epub 2014 May 19. Proc Natl Acad Sci U S A. 2014. PMID: 24843156 Free PMC article. - The features of mucosa-associated microbiota in primary sclerosing cholangitis.
Torres J, Bao X, Goel A, Colombel JF, Pekow J, Jabri B, Williams KM, Castillo A, Odin JA, Meckel K, Fasihuddin F, Peter I, Itzkowitz S, Hu J. Torres J, et al. Aliment Pharmacol Ther. 2016 Apr;43(7):790-801. doi: 10.1111/apt.13552. Epub 2016 Feb 9. Aliment Pharmacol Ther. 2016. PMID: 26857969 Free PMC article.
References
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A. et al.Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;5:119–124. doi: 10.1038/nature11582. - DOI - PMC - PubMed
- Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;5:120–123. doi: 10.1126/science.1224820. - DOI - PMC - PubMed
Publication types
Grants and funding
- K08 AI078942/AI/NIAID NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- R01 CA154426/CA/NCI NIH HHS/United States
- R01 DK092405/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources