Hepatic gene expression profiles differentiate presymptomatic patients with mild versus severe nonalcoholic fatty liver disease - PubMed (original) (raw)
Comparative Study
doi: 10.1002/hep.26661. Epub 2013 Dec 13.
Herbert Pang, Andrew Dellinger, Ayako Suzuki, Melanie E Garrett, Cynthia D Guy, Susan K Murphy, Allison E Ashley-Koch, Steve S Choi, Gregory A Michelotti, Daniel D Hampton, Yuping Chen, Hans L Tillmann, Michael A Hauser, Manal F Abdelmalek, Anna Mae Diehl
Affiliations
- PMID: 23913408
- PMCID: PMC3982589
- DOI: 10.1002/hep.26661
Comparative Study
Hepatic gene expression profiles differentiate presymptomatic patients with mild versus severe nonalcoholic fatty liver disease
Cynthia A Moylan et al. Hepatology. 2014 Feb.
Abstract
Clinicians rely upon the severity of liver fibrosis to segregate patients with well-compensated nonalcoholic fatty liver disease (NAFLD) into subpopulations at high- versus low-risk for eventual liver-related morbidity and mortality. We compared hepatic gene expression profiles in high- and low-risk NAFLD patients to identify processes that distinguish the two groups and hence might be novel biomarkers or treatment targets. Microarray analysis was used to characterize gene expression in percutaneous liver biopsies from low-risk, "mild" NAFLD patients (fibrosis stage 0-1; n = 40) and high-risk, "severe" NAFLD patients (fibrosis stage 3-4; n = 32). Findings were validated in a second, independent cohort and confirmed by real-time polymerase chain reaction and immunohistochemistry (IHC). As a group, patients at risk for bad NAFLD outcomes had significantly worse liver injury and more advanced fibrosis (severe NAFLD) than clinically indistinguishable NAFLD patients with a good prognosis (mild NAFLD). A 64-gene profile reproducibly differentiated severe NAFLD from mild NAFLD, and a 20-gene subset within this profile correlated with NAFLD severity, independent of other factors known to influence NAFLD progression. Multiple genes involved with tissue repair/regeneration and certain metabolism-related genes were induced in severe NAFLD. Ingenuity Pathway Analysis and IHC confirmed deregulation of metabolic and regenerative pathways in severe NAFLD and revealed overlap among the gene expression patterns of severe NAFLD, cardiovascular disease, and cancer.
Conclusion: By demonstrating specific metabolic and repair pathways that are differentially activated in livers with severe NAFLD, gene profiling identified novel targets that can be exploited to improve diagnosis and treatment of patients who are at greatest risk for NAFLD-related morbidity and mortality.
© 2013 by the American Association for the Study of Liver Diseases.
Conflict of interest statement
CONFLICT OF INTEREST
Potential competing Interests: None.
Figures
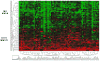
Figure 1. Hierarchical clustering analysis
Hierarchical clustering of the top 100 differentially expressed probes from the 72 NAFLD patients separated the samples into two main groups: mild NAFLD and severe NAFLD. Data are presented in heat map format in which patient samples are shown in rows and genes (probes) in columns. Red color corresponds to genes that are up-regulated in severe NAFLD as compared to the mean, and green color corresponds to genes that are down-regulated in severe NAFLD as compared to the mean.
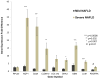
Figure 2. Quantitative RT-PCR of human NAFLD RNA samples
Bar graphs represent the fold difference in gene expression for mild NAFLD as compared to severe NAFLD. Dark bars represent the average gene expression in mild NAFLD from 7 unique patients while the light bars represent the average gene expression in severe NAFLD from 7 unique patients. RPL35 was used as the control gene for all analyses. Results are means ± SE.
Figure 3
Hepatic accumulation of markers of tissue repair and regeneration and liver progenitor cells is greater in severe NAFLD compared to mild NAFLD. Photomicrographs of SHH, GLI2, K7, SOX9 and α-SMA IHC in patients with mild and advanced NAFLD are shown (400x magnification). Liver sections stained for SHH (brown, arrowheads) show greater numbers of positive cells in severe NAFLD (B) compared to mild NAFLD (A). Liver sections double stained for K7 (brown) and GLI2 (red, nuclear, arrow head) reveals significantly higher grade of co-staining (arrows) in livers with severe NAFLD (D) than in mild NAFLD (C). Photomicrographs illustrate parallel accumulation of liver progenitor cells co-expressing GLI2 (red, nuclear) and SOX9 (blue, nuclear), along with α-SMA-positive myofibroblastic cells (brown) in severe NAFLD (F) compared to mild NAFLD (E). Many bile ductular cells in severe NAFLD (1000x magnification) express SOX9 (blue, nuclear, arrows); such cells generally co-express GLI2 (red, nuclear, arrow heads)(G). Figure and table summarizing the semi-quantitative immunohistochemistry results are shown in the Supplementary Materials and Methods.
Comment in
- NAFLD: Profiling NAFLD--liver gene expression and DNA methylation patterns to characterize disease severity.
Ray K. Ray K. Nat Rev Gastroenterol Hepatol. 2013 Oct;10(10):565. doi: 10.1038/nrgastro.2013.161. Epub 2013 Aug 20. Nat Rev Gastroenterol Hepatol. 2013. PMID: 23958603 No abstract available. - Reply: To PMID 23913408.
Moylan CA, Pang H, Michelotti G, Diehl AM. Moylan CA, et al. Hepatology. 2014 Oct;60(4):1445-6. doi: 10.1002/hep.27038. Epub 2014 Aug 13. Hepatology. 2014. PMID: 24493022 Free PMC article. No abstract available. - Normalization of a NAFLD gene expression signature.
Vinciguerra M. Vinciguerra M. Hepatology. 2014 Oct;60(4):1445. doi: 10.1002/hep.27042. Epub 2014 Aug 21. Hepatology. 2014. PMID: 24493162 No abstract available.
Similar articles
- Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease.
Murphy SK, Yang H, Moylan CA, Pang H, Dellinger A, Abdelmalek MF, Garrett ME, Ashley-Koch A, Suzuki A, Tillmann HL, Hauser MA, Diehl AM. Murphy SK, et al. Gastroenterology. 2013 Nov;145(5):1076-87. doi: 10.1053/j.gastro.2013.07.047. Epub 2013 Jul 31. Gastroenterology. 2013. PMID: 23916847 Free PMC article. - Serum coding and non-coding RNAs as biomarkers of NAFLD and fibrosis severity.
Di Mauro S, Scamporrino A, Petta S, Urbano F, Filippello A, Ragusa M, Di Martino MT, Scionti F, Grimaudo S, Pipitone RM, Privitera G, Di Pino A, Scicali R, Valenti L, Dongiovanni P, Fracanzani A, Rabuazzo AM, Craxì A, Purrello M, Purrello F, Piro S. Di Mauro S, et al. Liver Int. 2019 Sep;39(9):1742-1754. doi: 10.1111/liv.14167. Epub 2019 Jun 26. Liver Int. 2019. PMID: 31169972 Free PMC article. - Hepatic transcriptome signature correlated with HOMA-IR explains early nonalcoholic fatty liver disease pathogenesis.
Chatterjee A, Basu A, Das K, Singh P, Mondal D, Bhattacharya B, Roychoudhury S, Majumder PP, Chowdhury A, Basu P. Chatterjee A, et al. Ann Hepatol. 2020 Sep-Oct;19(5):472-481. doi: 10.1016/j.aohep.2020.06.009. Epub 2020 Jul 15. Ann Hepatol. 2020. PMID: 32682086 - Molecular Mechanisms: Connections between Nonalcoholic Fatty Liver Disease, Steatohepatitis and Hepatocellular Carcinoma.
Kanda T, Goto T, Hirotsu Y, Masuzaki R, Moriyama M, Omata M. Kanda T, et al. Int J Mol Sci. 2020 Feb 23;21(4):1525. doi: 10.3390/ijms21041525. Int J Mol Sci. 2020. PMID: 32102237 Free PMC article. Review. - Noninvasive Assessment of Liver Disease in Patients With Nonalcoholic Fatty Liver Disease.
Castera L, Friedrich-Rust M, Loomba R. Castera L, et al. Gastroenterology. 2019 Apr;156(5):1264-1281.e4. doi: 10.1053/j.gastro.2018.12.036. Epub 2019 Jan 18. Gastroenterology. 2019. PMID: 30660725 Free PMC article. Review.
Cited by
- Gene Expression Predicts Histological Severity and Reveals Distinct Molecular Profiles of Nonalcoholic Fatty Liver Disease.
Hoang SA, Oseini A, Feaver RE, Cole BK, Asgharpour A, Vincent R, Siddiqui M, Lawson MJ, Day NC, Taylor JM, Wamhoff BR, Mirshahi F, Contos MJ, Idowu M, Sanyal AJ. Hoang SA, et al. Sci Rep. 2019 Aug 29;9(1):12541. doi: 10.1038/s41598-019-48746-5. Sci Rep. 2019. PMID: 31467298 Free PMC article. - Hepatic Gene Expression Profiles Differentiate Steatotic and Non-steatotic Grafts in Liver Transplant Recipients.
Šeda O, Cahová M, Míková I, Šedová L, Daňková H, Heczková M, Brátová M, Ďásková N, Erhartová D, Čapek V, Chylíková B, Trunečka P. Šeda O, et al. Front Endocrinol (Lausanne). 2019 Apr 30;10:270. doi: 10.3389/fendo.2019.00270. eCollection 2019. Front Endocrinol (Lausanne). 2019. PMID: 31114547 Free PMC article. - NAFLD: Profiling NAFLD--liver gene expression and DNA methylation patterns to characterize disease severity.
Ray K. Ray K. Nat Rev Gastroenterol Hepatol. 2013 Oct;10(10):565. doi: 10.1038/nrgastro.2013.161. Epub 2013 Aug 20. Nat Rev Gastroenterol Hepatol. 2013. PMID: 23958603 No abstract available. - Hedgehog-YAP Signaling Pathway Regulates Glutaminolysis to Control Activation of Hepatic Stellate Cells.
Du K, Hyun J, Premont RT, Choi SS, Michelotti GA, Swiderska-Syn M, Dalton GD, Thelen E, Rizi BS, Jung Y, Diehl AM. Du K, et al. Gastroenterology. 2018 Apr;154(5):1465-1479.e13. doi: 10.1053/j.gastro.2017.12.022. Epub 2018 Jan 3. Gastroenterology. 2018. PMID: 29305935 Free PMC article. - Biomarkers and subtypes of deranged lipid metabolism in non-alcoholic fatty liver disease.
Mato JM, Alonso C, Noureddin M, Lu SC. Mato JM, et al. World J Gastroenterol. 2019 Jun 28;25(24):3009-3020. doi: 10.3748/wjg.v25.i24.3009. World J Gastroenterol. 2019. PMID: 31293337 Free PMC article. Review.
References
- Matteoni CA, Younossi ZM, Gramlich T, Boparai N, Liu YC, McCullough AJ. Nonalcoholic fatty liver disease: a spectrum of clinical and pathological severity. Gastroenterology. 1999;116:1413–1419. - PubMed
- Rafiq N, Bai C, Fang Y, Srishord M, McCullough A, Gramlich T, Younossi ZM. Long-term follow-up of patients with nonalcoholic fatty liver. Clin Gastroenterol Hepatol. 2009;7:234–238. - PubMed
- Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. - PubMed
- Younossi ZM, Stepanova M, Rafiq N, Makhlouf H, Younoszai Z, Agrawal R, Goodman Z. Pathologic criteria for nonalcoholic steatohepatitis: interprotocol agreement and ability to predict liver-related mortality. Hepatology. 2011;53:1874–1882. - PubMed
Publication types
MeSH terms
Grants and funding
- K23 DK062116/DK/NIDDK NIH HHS/United States
- U01-DK57149/DK/NIDDK NIH HHS/United States
- U01 DK057149/DK/NIDDK NIH HHS/United States
- 5RC2 AA019399/AA/NIAAA NIH HHS/United States
- T32 DK007568/DK/NIDDK NIH HHS/United States
- RC2 AA019399/AA/NIAAA NIH HHS/United States
- K23-DK062116/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials