Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex - PubMed (original) (raw)
Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex
Derya Dönertas et al. Genes Dev. 2013.
Abstract
The PIWI-interacting RNA (piRNA) pathway is a small RNA silencing system that keeps selfish genetic elements such as transposons under control in animal gonads. Several lines of evidence indicate that nuclear PIWI family proteins guide transcriptional silencing of their targets, yet the composition of the underlying silencing complex is unknown. Here we demonstrate that the double CHHC zinc finger protein gametocyte-specific factor 1 (Gtsf1) is an essential factor for Piwi-mediated transcriptional repression in Drosophila. Cells lacking Gtsf1 contain nuclear Piwi loaded with piRNAs, yet Piwi's silencing capacity is ablated. Gtsf1 interacts directly with a small subpool of nuclear Piwi, and loss of Gtsf1 phenocopies loss of Piwi in terms of deregulation of transposons, loss of H3K9 trimethylation (H3K9me3) marks at euchromatic transposon insertions, and deregulation of genes in proximity to repressed transposons. We propose that only a small fraction of nuclear Piwi is actively engaged in target silencing and that Gtsf1 is an essential component of the underlying Piwi-centered silencing complex.
Keywords: Gtsf1; Piwi; heterochromatin formation; piRNA pathway; transcriptional silencing; transposon control.
Figures
Figure 1.
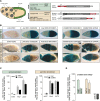
CG3893 is required for transposon silencing in ovarian somatic and germline cells. (A) Cartoon of a Drosophila ovariole, with follicle cells (soma) in green and nurse cells and oocyte (germline) in beige. To the right, lacZ sensors that report derepression of soma-specific (gypsy) or germline-specific (Burdock and HeT-A) TEs. For the soma reporter, _tj_-Gal4, _gypsy_-lacZ flies carrying a restrictive flamenco background were crossed to VDRC lines of interest (SKD). For the germline reporters, target gene knockdown was performed by either MTD-Gal4>UAS-shRNA or NGT-Gal4>UAS-Dcr-2/UAS-VDRC (GLKD). Portions of the respective reporters targeted by piRNAs are depicted in red. (B) Shown are β-Gal stainings of egg chambers expressing gypsy (top), Burdock (middle), or HeT-A (bottom) reporters. w[1118] flies (control) or indicated VDRC/shRNA lines for piwi, mael, or CG3893 were used for the analysis. (C) Displayed are fold changes in steady-state RNA levels of indicated TEs and genes in mael or CG3893 knockdown ovaries in log10 scale (soma, left panel; germline, right panel). Values are averages of three biological replicates (error bars indicate SD) and were normalized to control knockdowns via levels of the rp49 housekeeping gene. (D) Displayed are fold changes in steady-state RNA levels of indicated TEs ([green] soma-dominant; [beige] germline dominant) and genes in CG3893[GS12962] homozygous versus heterozygous ovaries in log10 scale.
Figure 2.
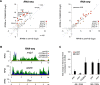
CG3893 is a core member of the somatic piRNA pathway. (A) Scatter plot of RNA-seq reads per kilobase per million mapped reads (RPKM) values (log2; ribozero OSC RNA) for annotated Drosophila melanogaster TEs (n = 125) in control knockdown (control-KD) versus CG3893 knockdown (_CG3893_-KD) (left) and piwi knockdown (_piwi_-KD) versus CG3893 knockdown (_CG3893_-KD) (right). Four TE groups are color-coded according to the classification in Sienski et al. (2012). (Red and orange) Piwi-repressed; (gray) nonrepressed; (open circles) nonexpressed. (B) Density profiles of normalized RNA-seq reads mapping to mdg1, gypsy (group I), or Doc (group III). The orange profile indicates the control knockdown (_GFP_-48h-KD) for the CG3893 knockdown (_CG3893_-KD; blue); the red profile indicates the control knockdown (_GFP_-96h-KD) for the _piwi_-knockdown (_piwi_-KD; green). (C) Time-course experiment showing fold changes in steady-state RNA levels of mdg1 (black bars) and act5C (red bars) after 48 h or 96 h of RNAi for the indicated genes in OSCs.
Figure 3.
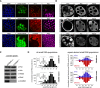
CG3893 is not required for piRNA biogenesis. (A) Confocal sections of follicular epithelia stained for Piwi (magenta), DNA (DAPI; blue), and the RNAi target gene (red) in which _armi_-RNAi, _mael_-RNAi, or _CG3893_-RNAi has been clonally activated (marked by GFP; boundary indicated by dashed yellow line). (B) Confocal sections of egg chambers with GLKDs of Piwi or CG3893 compared with a w[1118] cross (control), stained for Piwi, Aub, or AGO3. (C) Western blots showing protein levels of Piwi, Aub, AGO3, Mael, or CG3893 in CG3893[GS12962] heterozygous or homozygous ovaries. (D) To the left, length profiles of all small RNAs (normalized to 1 million miRNAs; small insets) isolated from ovaries of CG3893[GS12962] heterozygous or homozygous flies are shown (small RNAs mapping to rRNA, tRNAs, or snoRNAs have been computationally removed; siRNA and piRNA populations are indicated). Panels to the right show length profiles of repeat-derived ovarian small RNAs (normalized to 1 million miRNAs) from CG3893[GS12962] heterozygous or homozygous flies. (Red) Antisense; (blue) sense.
Figure 4.
CG3893 physically interacts with Piwi, and its nuclear localization depends on Piwi. (A, top panels) Confocal sections of an egg chamber expressing GFP-CG3893 from a genomic rescue construct stained for DNA (DAPI) and analyzed for GFP fluorescence. (Bottom panels) Confocal sections of egg chambers stained for CG3893 in control and CG3893 GLKDs. (B) SDS-PAGE of a CG3893 and IgG control immunoprecipitation (IP) from nuclear OSC lysate stained with silver. A molecular weight marker is indicated to the left, and CG3893 and Piwi bands are indicated. (C) Western blots showing co-IP of endogenous CG3893 with endogenous Piwi from nuclear OSC lysates. Rabbit IgG and mouse IgG were used as control immunoprecipitations for Piwi and CG3893, respectively. (D) Confocal images of a wild-type egg chamber stained for Piwi (top) and CG3893 (bottom). Arrowheads indicate areas with pronounced colocalization. (E) Confocal images of egg chambers stained for Piwi (top panels) and CG3893 (bottom panels). The target of the respective GLKD is indicated at the top. (F) Confocal images of egg chambers expressing GFP-tagged N-terminally truncated Piwi (top panels) stained for CG3893 (bottom panels). The left panels show ovaries with a wild-type piwi background, and the right panels show ovaries with a piwi[1]/[2] background.
Figure 5.
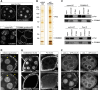
A C-terminal peptide in CG3893/Gtsf1 interacts with Piwi. (A) Cartoon of the CG3893/Gtsf1 protein showing the N-terminal zinc fingers and the unstructured C-terminal tail. Shown is the highly conserved amino acid stretch (residues 83–115) with negatively charged amino acids (blue) and the highly conserved aromatic W89 and Y98 residues. (B) Shown are derepression levels for the mdg1 and gypsy TEs upon CG3893 knockdown with simultaneous transfection of plasmids for the expression of siRNA-resistant CG3893 versions with indicated mutations. (ZnF) Zinc finger mutations; (ΔC) deletion of C-tail. (C) Shown are immunoprecipitation experiments (anti-Gtsf1 and anti-Piwi) from OSC nuclear lysate (amounts of total protein indicated) and pull-down experiments with indicated Gtsf1 peptides (residues 83–115) probed for Piwi (top blot) and Gtsf1 (bottom blot). (D) SDS-PAGE of Gtsf1 peptide (83–115) pull-downs (WY mutant, left; wild type, right) from nuclear OSC lysate stained with silver. A molecular weight marker is indicated to the left, and the Piwi band confirmed by mass spectrometry is marked. (E) Shown are peptide pull-down experiments with indicated Gtsf1 peptides where peptide amounts (1×–10×) have been varied. Input was OSC nuclear lysate, and eluates were analyzed by Western blot against Piwi (top), Gtsf1 (middle), and tubulin (bottom).
Figure 6.
CG3893/Gtsf1 is required for Piwi-dependent H3K9me3 domains and _trans_-silencing of individual TE insertions. (A) Transcriptional activity (Pol II ChIP-seq), H3K9me3 signal (H3K9me3 ChIP-seq), and steady-state RNA levels (RNA-seq) at the CG15828 locus are shown for OSCs with the indicated knockdowns (input tracks for the respective ChIP experiments are in gray). An OSC-specific mdg1 insertion is indicated at the top; the transcriptional start site (TSS) of CG15828 is indicated with a dashed line. (B,C) Heat maps showing normalized signal of H3K9me3 (B) or Pol II occupancy (C) in windows of 50 kb centered on summits of euchromatic H3K9me3 peaks (n = 466) in control knockdown, piwi knockdown (_piwi_-KD), or CG3893/Gtsf1 knockdown (_CG3893/Gtsf1_-KD). For the H3K9me3 heat map, peaks were sorted according to the loss of signal in the 1-kb window around the peak summit in _piwi_-KD cells; for the Pol II heat map, peaks were sorted according to gain of signal in the 1-kb window around the peak summit in piwi knockdown cells. The H3K9me3 heat map was split into five equally sized bins as in Sienski et al. (2012), and their average signals are shown in Supplemental Fig. S8A. (D) Box plot showing the fold changes (log2) in RNA-seq RPKM values for the set of expressed genes (RPKM > 5 in any of the libraries) with a nearby insertion of a group I TE in sense orientation (red) or a nearby insertion of a group III TE in sense orientation (gray) in piwi knockdown (_piwi_-KD; left) or CG3893/Gtsf1 knockdown (_CG3893/Gtsf1_-KD; right) OSCs. _P_-value was calculated according to Wilcoxon rank-sum test.
Similar articles
- DmGTSF1 is necessary for Piwi-piRISC-mediated transcriptional transposon silencing in the Drosophila ovary.
Ohtani H, Iwasaki YW, Shibuya A, Siomi H, Siomi MC, Saito K. Ohtani H, et al. Genes Dev. 2013 Aug 1;27(15):1656-61. doi: 10.1101/gad.221515.113. Genes Dev. 2013. PMID: 23913921 Free PMC article. - Silencio/CG9754 connects the Piwi-piRNA complex to the cellular heterochromatin machinery.
Sienski G, Batki J, Senti KA, Dönertas D, Tirian L, Meixner K, Brennecke J. Sienski G, et al. Genes Dev. 2015 Nov 1;29(21):2258-71. doi: 10.1101/gad.271908.115. Epub 2015 Oct 22. Genes Dev. 2015. PMID: 26494711 Free PMC article. - Asterix/Gtsf1 links tRNAs and piRNA silencing of retrotransposons.
Ipsaro JJ, O'Brien PA, Bhattacharya S, Palmer AG 3rd, Joshua-Tor L. Ipsaro JJ, et al. Cell Rep. 2021 Mar 30;34(13):108914. doi: 10.1016/j.celrep.2021.108914. Cell Rep. 2021. PMID: 33789107 Free PMC article. - To export, or not to export: how nuclear export factor variants resolve Piwi's dilemma.
Wang S, Lu X, Qiu D, Yu Y. Wang S, et al. Biochem Soc Trans. 2021 Nov 1;49(5):2073-2079. doi: 10.1042/BST20201171. Biochem Soc Trans. 2021. PMID: 34643228 Review. - The piRNA pathway in Drosophila ovarian germ and somatic cells.
Sato K, Siomi MC. Sato K, et al. Proc Jpn Acad Ser B Phys Biol Sci. 2020;96(1):32-42. doi: 10.2183/pjab.96.003. Proc Jpn Acad Ser B Phys Biol Sci. 2020. PMID: 31932527 Free PMC article. Review.
Cited by
- Mouse GTSF1 is an essential factor for secondary piRNA biogenesis.
Yoshimura T, Watanabe T, Kuramochi-Miyagawa S, Takemoto N, Shiromoto Y, Kudo A, Kanai-Azuma M, Tashiro F, Miyazaki S, Katanaya A, Chuma S, Miyazaki JI. Yoshimura T, et al. EMBO Rep. 2018 Apr;19(4):e42054. doi: 10.15252/embr.201642054. Epub 2018 Feb 7. EMBO Rep. 2018. PMID: 29437694 Free PMC article. - Transposable element dynamics and PIWI regulation impacts lncRNA and gene expression diversity in Drosophila ovarian cell cultures.
Sytnikova YA, Rahman R, Chirn GW, Clark JP, Lau NC. Sytnikova YA, et al. Genome Res. 2014 Dec;24(12):1977-90. doi: 10.1101/gr.178129.114. Epub 2014 Sep 29. Genome Res. 2014. PMID: 25267525 Free PMC article. - Impact of nuclear Piwi elimination on chromatin state in Drosophila melanogaster ovaries.
Klenov MS, Lavrov SA, Korbut AP, Stolyarenko AD, Yakushev EY, Reuter M, Pillai RS, Gvozdev VA. Klenov MS, et al. Nucleic Acids Res. 2014 Jun;42(10):6208-18. doi: 10.1093/nar/gku268. Epub 2014 Apr 29. Nucleic Acids Res. 2014. PMID: 24782529 Free PMC article. - Small RNA profiling and characterization of piRNA clusters in the adult testes of the common marmoset, a model primate.
Hirano T, Iwasaki YW, Lin ZY, Imamura M, Seki NM, Sasaki E, Saito K, Okano H, Siomi MC, Siomi H. Hirano T, et al. RNA. 2014 Aug;20(8):1223-37. doi: 10.1261/rna.045310.114. Epub 2014 Jun 9. RNA. 2014. PMID: 24914035 Free PMC article. - Zucchini-dependent piRNA processing is triggered by recruitment to the cytoplasmic processing machinery.
Rogers AK, Situ K, Perkins EM, Toth KF. Rogers AK, et al. Genes Dev. 2017 Sep 15;31(18):1858-1869. doi: 10.1101/gad.303214.117. Epub 2017 Oct 11. Genes Dev. 2017. PMID: 29021243 Free PMC article.
References
- Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT 2005. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell 17: 103–112 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous