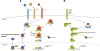The complexity of NF-κB signaling in inflammation and cancer - PubMed (original) (raw)
Review
The complexity of NF-κB signaling in inflammation and cancer
Bastian Hoesel et al. Mol Cancer. 2013.
Abstract
The NF-κB family of transcription factors has an essential role in inflammation and innate immunity. Furthermore, NF-κB is increasingly recognized as a crucial player in many steps of cancer initiation and progression. During these latter processes NF-κB cooperates with multiple other signaling molecules and pathways. Prominent nodes of crosstalk are mediated by other transcription factors such as STAT3 and p53 or the ETS related gene ERG. These transcription factors either directly interact with NF-κB subunits or affect NF-κB target genes. Crosstalk can also occur through different kinases, such as GSK3-β, p38, or PI3K, which modulate NF-κB transcriptional activity or affect upstream signaling pathways. Other classes of molecules that act as nodes of crosstalk are reactive oxygen species and miRNAs. In this review, we provide an overview of the most relevant modes of crosstalk and cooperativity between NF-κB and other signaling molecules during inflammation and cancer.
Figures
Figure 1
Members of the NF-κB signaling pathway and the IκB kinase-complex. (A) The five members of the NF-κB family of proteins: RelA (p65), RelB, c-Rel,NF-κB1 (p105), and NF-κB2 (p100). p105 and p100 are processed to their shorter forms p50 and p52, respectively. All members of the NF-κB family harbor an N-terminal Rel homology domain (RHD), which mediates DNA contact and homo- and heterodimerization. Three family members (RelA, RelB and c-Rel) contain C-terminal transactivation domains (TAs), which are essential for transcriptional activity. (B) The IκB family of proteins consists of four members: IκBα, IκBβ, IκBϵ and BCL-3. These proteins are characterized by the presence of ankyrin (ANK) repeats, which mediate binding of IκBs to the NF-κB family of proteins. Based on the presence of ankyrin repeats, p100 and p105 can also be included into the IκB family – as their DNA-binding RHD domain is covalently linked to an IκB-like inhibitory domain. In addition to the ANK repeats IκBα and IκBβ contain PEST domains, which are enriched in proline, glutamate, serine and threonine and are required for constitutive turnover. BCL-3 differs from other IκB family members by containing TA domains, which mediate transcriptional activity when BCL-3 is associated with NF-κB dimers that bind to DNA. (C) The three most important members of IκB kinase (IKK) complex: NF-κB Essential Modulator (NEMO or IKKγ), IκB kinase α, (IKKα or IKK1) and IκB kinase β (IKKβ or IKK2). Further abbreviations: leucin-zipper-like motif (LZ), death domain (DD), coiled-coil domain (CC), zinc-finger domain (ZF), helix-loop-helix domain (HLH), NEMO-binding domain (NBD). It is important to note that the total number of amino acids of protein as well as the start and end of some domains can differ between publications and databases.
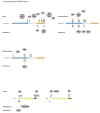
Figure 2
The canonical, non-canonical and the atypical NF-κB signaling pathway. (A) In the canonical NF-κB signaling pathway lipopolysaccharides (LPS), tumor necrosis factor α (TNFα) orinterleukin-1 (IL-1) activate Toll-like receptors (TLRs), tumor necrosis factor receptor (TNFR) and interleukin-1 receptor (IL-1R), respectively. Through a variety of adapter proteins and signaling kinases this leads to an activation of IKKβ in the IKK complex, which can then phosphorylate IκBα on Serine residues S32 and S36. This phosphorylation is a prerequisite for its subsequent polyubiquitination, which in turn results in proteasomal degradation of IκBα. NF-κB homo- or heterodimers can then translocate to nucleus and activate target gene transcription. (B) In the non-canonical NF-κB signaling pathway, activation of B-cell activation factor (BAFFR), CD40, receptor activator for nuclear factor kappa B (RANK) or lymphtoxin β-receptor (LTβR), leads to activation of IKKα by the NF-κB-inducing kinase (NIK). IKKα can the phosphorylate p100 on serine residues S866 and S870. This phosphorylation leads to polyubiquitination of p100 and its subsequent proteasomal processing to p52.p52-RelB heterodimers can then activate transcription of target genes. (C) In the atypical NF-κB signaling pathway, genotoxic stress leads to a translocation of NEMO to the nucleus where it is sumoylated and subsequently ubiquitinated. This process is mediated by the ataxia telangiectasia mutated (ATM) checkpoint kinase. NEMO and ATM can then return to the cytosol where they activate IKKβ.
Figure 3
Post-translational modifications of RelA, IκBα and IκBβ. Phosphorylations, acetylations and methylations of RelA are shown, as well as phosphorylations, ubiquitination and sumoylation of IkBα and IkB.
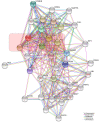
Figure 4
Network of NF-κB interactors. Evidence view of the STRING database output depicting functional and physical interactors of the NF-κB proteins, RelA, Rel (c-Rel), RelB, NFKB1 and NFKB2 obtained from:
. The five NF-κB proteins are highlighted in red.
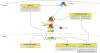
Figure 5
Crosstalk of the canonical NF-κB pathway with other signaling processes. (A) Many different kinases can phosphorylate and activate the IKKα and IKKβ subunits of the IKK complex or can enhance NF-κB transcriptional activity. Important examples are glycogen synthase kinase 3β (GSK3β), Protein Kinase B (PKB or Akt), Protein Kinase R (PKR), Protein Kinase C (PKC), Mitogen-Activated Type 3-Protein Kinase 7 (MAP3K7 or TAK1), p38 MAP Kinases or c-Jun N-terminal kinases (JNKs). (B) Various transcription factors such as p53, Ets Related Gene (ERG) or Signal Transducer and Activator of Transcription 3 (STAT3) can influence the transcriptional activity of NF-κB or directly activate transcription of NF-κB target genes. (C) microRNAs (miRNAs) can be target genes of the NF-κB signaling pathways or can affect the expression of NF-κB family members or effector molecules of the NF-κB activation pathway. (D) Prominent target genes of the NF-κB signaling pathway include anti-apoptotic genes as the Baculoviral IAP repeat-containing proteins (BIRCs or cIAPs) and the B-cell lymphoma 2 gene (Bcl-2), cytokines such as Interleukin-1 (IL-1), IL-6, IL-8 and chemokine (C-C motif) ligand 2 (CCL2), adhesion factors including the Vascular Cell Adhesion Molecule 1 (VCAM-1) and the Intercellular Cell Adhesion Molecule 1 (ICAM-1). (E) Another layer of complexity of NF-κB signaling are positive and negative feedback mechanism. Examples for positive feedback molecules are the X-linked inhibitor of apoptosis protein (XIAP) as well as TNFα or IL-1. Important negative feedback circuits are generated by the NF-κB target genes IκBα, Cylindromatosis (CYLD) or A20.
Similar articles
- Antagonistic crosstalk between NF-κB and SIRT1 in the regulation of inflammation and metabolic disorders.
Kauppinen A, Suuronen T, Ojala J, Kaarniranta K, Salminen A. Kauppinen A, et al. Cell Signal. 2013 Oct;25(10):1939-48. doi: 10.1016/j.cellsig.2013.06.007. Epub 2013 Jun 11. Cell Signal. 2013. PMID: 23770291 Review. - Regulation of the MIR155 host gene in physiological and pathological processes.
Elton TS, Selemon H, Elton SM, Parinandi NL. Elton TS, et al. Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review. - Regulation of Nuclear Factor-KappaB (NF-κB) signaling pathway by non-coding RNAs in cancer: Inhibiting or promoting carcinogenesis?
Mirzaei S, Zarrabi A, Hashemi F, Zabolian A, Saleki H, Ranjbar A, Seyed Saleh SH, Bagherian M, Sharifzadeh SO, Hushmandi K, Liskova A, Kubatka P, Makvandi P, Tergaonkar V, Kumar AP, Ashrafizadeh M, Sethi G. Mirzaei S, et al. Cancer Lett. 2021 Jul 1;509:63-80. doi: 10.1016/j.canlet.2021.03.025. Epub 2021 Apr 7. Cancer Lett. 2021. PMID: 33838282 Review. - MicroRNAs in NF-kappaB signaling.
Ma X, Becker Buscaglia LE, Barker JR, Li Y. Ma X, et al. J Mol Cell Biol. 2011 Jun;3(3):159-66. doi: 10.1093/jmcb/mjr007. Epub 2011 Apr 18. J Mol Cell Biol. 2011. PMID: 21502305 Free PMC article. Review. - Differential expression of miRNAs regulating NF-κB and STAT3 crosstalk during colitis-associated tumorigenesis.
El-Daly SM, Omara EA, Hussein J, Youness ER, El-Khayat Z. El-Daly SM, et al. Mol Cell Probes. 2019 Oct;47:101442. doi: 10.1016/j.mcp.2019.101442. Epub 2019 Aug 31. Mol Cell Probes. 2019. PMID: 31479716
Cited by
- Exploring the Phytochemistry, Signaling Pathways, and Mechanisms of Action of Tanacetum parthenium (L.) Sch.Bip.: A Comprehensive Literature Review.
Kashkooe A, Jalali A, Zarshenas MM, Hamedi A. Kashkooe A, et al. Biomedicines. 2024 Oct 10;12(10):2297. doi: 10.3390/biomedicines12102297. Biomedicines. 2024. PMID: 39457613 Free PMC article. Review. - Protective effects of N-acetylcysteine in concanavalin A-induced hepatitis in mice.
Wang C, Xia Y, Zheng Y, Dai W, Wang F, Chen K, Li J, Li S, Zhu R, Yang J, Yin Q, Zhang H, Wang J, Lu J, Zhou Y, Guo C. Wang C, et al. Mediators Inflamm. 2015;2015:189785. doi: 10.1155/2015/189785. Epub 2015 Mar 2. Mediators Inflamm. 2015. PMID: 25821351 Free PMC article. - Chidamide induces apoptosis in DLBCL cells by suppressing the HDACs/STAT3/Bcl‑2 pathway.
Zhang H, Chi F, Qin K, Mu X, Wang L, Yang B, Wang Y, Bai M, Li Z, Su L, Yu B. Zhang H, et al. Mol Med Rep. 2021 May;23(5):308. doi: 10.3892/mmr.2021.11947. Epub 2021 Mar 2. Mol Med Rep. 2021. PMID: 33649847 Free PMC article. - miR-9 alleviated the inflammatory response and apoptosis in caerulein-induced acute pancreatitis by regulating FGF10 and the NF-κB signaling pathway.
Shen Y, Xue C, You G, Liu C. Shen Y, et al. Exp Ther Med. 2021 Aug;22(2):795. doi: 10.3892/etm.2021.10227. Epub 2021 May 25. Exp Ther Med. 2021. PMID: 34093751 Free PMC article. - To be an ally or an adversary in bladder cancer: the NF-κB story has not unfolded.
Mukherjee N, Houston TJ, Cardenas E, Ghosh R. Mukherjee N, et al. Carcinogenesis. 2015 Mar;36(3):299-306. doi: 10.1093/carcin/bgu321. Epub 2014 Dec 27. Carcinogenesis. 2015. PMID: 25543121 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous