Nav1.1 haploinsufficiency in excitatory neurons ameliorates seizure-associated sudden death in a mouse model of Dravet syndrome - PubMed (original) (raw)
. 2013 Dec 1;22(23):4784-804.
doi: 10.1093/hmg/ddt331. Epub 2013 Aug 6.
Affiliations
- PMID: 23922229
- PMCID: PMC3820136
- DOI: 10.1093/hmg/ddt331
Nav1.1 haploinsufficiency in excitatory neurons ameliorates seizure-associated sudden death in a mouse model of Dravet syndrome
Ikuo Ogiwara et al. Hum Mol Genet. 2013.
Abstract
Dravet syndrome is a severe epileptic encephalopathy mainly caused by heterozygous mutations in the SCN1A gene encoding a voltage-gated sodium channel Nav1.1. We previously reported dense localization of Nav1.1 in parvalbumin (PV)-positive inhibitory interneurons in mice and abnormal firing of those neurons in Nav1.1-deficient mice. In the present study, we investigated the physiologic consequence of selective Nav1.1 deletion in mouse global inhibitory neurons, forebrain excitatory neurons or PV cells, using vesicular GABA transporter (VGAT)-Cre, empty spiracles homolog 1 (Emx1)-Cre or PV-Cre recombinase drivers. We show that selective Nav1.1 deletion using VGAT-Cre causes epileptic seizures and premature death that are unexpectedly more severe than those observed in constitutive Nav1.1-deficient mice. Nav1.1 deletion using Emx1-Cre does not cause any noticeable abnormalities in mice; however, the severe lethality observed with VGAT-Cre-driven Nav1.1 deletion is rescued by additional Nav1.1 deletion using Emx1-Cre. In addition to predominant expression in PV interneurons, we detected Nav1.1 in subpopulations of excitatory neurons, including entorhino-hippocampal projection neurons, a subpopulation of neocortical layer V excitatory neurons, and thalamo-cortical projection neurons. We further show that even minimal selective Nav1.1 deletion, using PV-Cre, is sufficient to cause spontaneous epileptic seizures and ataxia in mice. Overall, our results indicate that functional impairment of PV inhibitory neurons with Nav1.1 haploinsufficiency contributes to the epileptic pathology of Dravet syndrome, and show for the first time that Nav1.1 haploinsufficiency in excitatory neurons has an ameliorating effect on the pathology.
Figures
Figure 1.
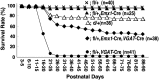
Generation of a floxed Scn1a allele in mice. (A) Verification of floxed and deleted exon 7 alleles using PCR analysis of genomic DNA. Primer positions and PCR product sizes are indicated. (B) Western blot analysis of brain membrane proteins prepared from 5-month-old wild-type and homozygous floxed Scn1a (_Scn1a_fl/fl) mice, using anti-Nav1.1 antibody. β-Tubulin was used as an internal control, and two independent assays were performed. (C) Western blot analysis of brain membrane proteins prepared from P14.5 Scn1a+/+, _Scn1a_d/+ and _Scn1a_d/d littermates, using anti-Nav1.1 antibody. β-Tubulin was used as an internal control, and two independent assays were performed. (D and E) Representative interictal (D) and ictal (E) ECoG recordings from _Scn1a_d/d mice. ECoG recordings were performed with P14–16 _Scn1a_d/d mice and Scn1a+/+ controls (n = 3, each group). (F) Survival curves of P3 Scn1a+/+, _Scn1a_d/+ and _Scn1a_d/d littermates. +, wild-type allele; fl, floxed allele; d, deleted allele.
Figure 2.
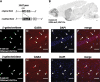
Generation of _VGAT_-Cre transgenic mice. (A) Schematic representation of the DNA construct used to generate the _VGAT_-Cre transgenic mouse line. (B) A representative parasagittal section from 8-week-old Rosa26-LacZ,_VGAT_-Cre brain, stained with anti-β-galactosidase antibody. Scale bar: 300 µm. (C–J) Representative images of immunofluorescence histochemistry in the hippocampus (C–F) and neocortical layer II/III (G–J) in 8-week-old Rosa26-LacZ,_VGAT_-Cre mice, stained with anti-β-galactosidase antibody (C and G; green), anti-GABA antibody (D and H; red) and DAPI (E and I; blue), with respective merged images (F and J). Arrowheads indicate cells double-labeled with β-galactosidase and GABA. Note that cells immunopositive for β-galactosidase (indicative of Cre-loxP recombination in the Rosa26-LacZ allele) were usually immunopositive for GABA also. o, stratum oriens; p, stratum pyramidale; r, stratum radiatum. Scale bars: 50 µm.
Figure 3.
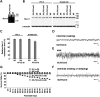
Selective Nav1.1 deletion in mouse inhibitory neurons, using _VGAT_-Cre results in severe epileptic seizures and high lethality. (A) Verification of _VGAT_-Cre-dependent Scn1a gene deletion in the floxed allele in P21.5 _Scn1a_fl/+,_VGAT_-Cre brain. PCR analysis of brain genomic DNA isolated from P21.5 _Scn1a_fl/+, _VGAT_-Cre and _Scn1a_fl/fl mice was performed. (B and C) Verification of _VGAT_-Cre-dependent Nav1.1 deletion by semi-quantitative western blot analysis of brain membrane proteins prepared from P21.5 _Scn1a_fl/+, _VGAT_-Cre mice and _Scn1a_fl/fl littermates, using anti-Nav1.1 antibody (B). β-Tubulin was used as an internal control. Mean Nav1.1 expression levels in _Scn1a_fl/+, _VGAT_-Cre mice are represented as percentages, relative to Nav1.1 in _Scn1a_fl/+ littermates (100%; C). (D and E) Representative ECoG of a lethal seizure in a P18–21 _Scn1a_fl/+,_VGAT_-Cre mouse. An arrow indicates onset of the epileptiform discharge. ECoG recordings were performed with P18–21 _Scn1a_fl/+,_VGAT_-Cre and _Scn1a_fl/+ control mice (n = 3, each group). (F) Survival curves of P3 _Scn1a_fl/+, _Scn1a_d/+ (reprinted from Fig. 1F for comparison), _Scn1a_fl/+,_VGAT_-Cre and _Scn1a_fl/fl,_VGAT_-Cre mice. Note that all _Scn1a_fl/fl,_VGAT_-Cre mice died before P15, and that all but one _Scn1a_fl/+,_VGAT_-Cre mice died before P35. (G) Verification of _VGAT_-Cre-dependent Scn1a gene deletion in the floxed allele in P12.5 _Scn1a_fl/fl,_VGAT_-Cre brain by PCR analysis. (H and I) Verification of _VGAT_-Cre-dependent Nav1.1 deletion by semi-quantitative western blot analysis of brain membrane proteins prepared from P12.5 _Scn1a_fl/fl, _VGAT_-Cre mice and age-matched _Scn1a_fl/fl controls, using anti-Nav1.1 antibody. Two independent assays were performed. Error bars represent SEM. *P < 0.05, **P < 0.01. +, wild-type allele; fl, floxed allele; d, deleted allele.
Figure 4.
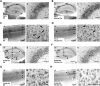
Nav1.1 immunosignals are significantly reduced in inhibitory neurons of _Scn1a_fl/fl,_VGAT_-Cre mice. (A–D) Representative parasagittal sections of P12.5 _Scn1a_fl/fl (A) and _Scn1a_fl/fl,_VGAT_-Cre (B) hippocampi, and P12.5 _Scn1a_fl/fl (C) and _Scn1a_fl/fl,_VGAT_-Cre (D) neocortices, stained with anti-Nav1.1 antibody. Higher magnification images outlined in (a) are shown in (b). An arrowhead indicates an Nav1.1-immunoreactive proximal neurite oriented toward pial surface, which putatively corresponds to an AIS of a PV cell (15,36) (see also Fig. 9O). Scale bars: (a) 400 µm; (b) 40 µm. DG, dentate gyrus; o, stratum oriens; p, stratum pyramidale; l, stratum lucidum. (E and F) Representative parasagittal sections of P12.5 _Scn1a_fl/fl (E) and _Scn1a_fl/fl, _VGAT_-Cre (F) cerebellum stained with anti-Nav1.1 antibody. Higher magnification images outlined in (a) are shown in (b) and (d). Higher magnification images outlined in (b) are shown in (c). Arrowheads indicate putative Nav1.1-immunoreactive AISs in Purkinje cells. Arrows indicate Nav1.1-immunoreactive axons of basket cells. Scale bars: (a) 400 µm; (b and c) 40 µm; (d) 80 µm. M, molecular cell layer; P, Purkinje cell layer; G, granule cell layer; CbN, cerebellar nuclei. Nuclear immunosignals are non-specific (15). Images are oriented from the pial surface (top) to the callosal (bottom), and from rostral (left) to caudal (right). fl, floxed allele.
Figure 5.
Selective Nav1.1 deletion in forebrain excitatory neurons by _Emx1_-Cre recombination did not cause any observable abnormality in mice. (A) Verification of _Emx1_-Cre-dependent deletion of the floxed Scn1a gene in the 8-week-old _Scn1a_fl/fl, _Emx1_-Cre brain by PCR analysis. (B and C) Verification of _Emx1_-Cre-dependent Nav1.1 deletion by semi-quantitative western blot analysis of brain membrane proteins prepared from P21.5 and 8-week-old _Scn1a_fl/fl, _Emx1_-Cre and _Scn1a_fl/fl littermates, using anti-Nav1.1 antibody. (D) Survival curves of P3 _Scn1a_fl/fl and _Scn1a_fl/fl, _Emx1_-Cre littermates. In both genotypes, a few mice died of unknown reasons. +, wild-type allele; fl, floxed allele. *P < 0.05, ***P < 0.001.
Figure 6.
Nav1.1 immunosignals in forebrain excitatory neurons are reduced in _Scn1a_fl/fl, _Emx1_-Cre mice. (A and B) Representative parasagittal sections of P21.5 _Scn1a_fl/fl (A) and _Scn1a_fl/fl, _Emx1_-Cre (B) hippocampi stained with anti-Nav1.1 antibody. Higher magnification images outlined in (a) are shown in (b). Scale bars: (a) 400 µm; (b) 40 µm. DG, dentate gyrus; o, stratum oriens; p, stratum pyramidale; l, stratum lucidum. (C and D) Representative parasagittal sections of P21.5 _Scn1a_fl/fl (C) and _Scn1a_fl/fl, _Emx1_-Cre (D) neocortices, stained with anti-Nav1.1 antibody. Higher magnification images outlined in (a) are shown in (b). Scale bars: (a) 400 µm; (b) 40 µm. Arrows indicate Nav1.1-immunoreactive neurites oriented toward the pial surface, which putatively correspond to AISs of neocortical PV cells (15,36) (see also Fig. 9O). Double arrowheads indicate Nav1.1-immunoreactive proximal neurites oriented toward the ventricular surface in neocortical layer V. (E–H) Representative parasagittal sections of 8-week-old _Scn1a_fl/fl (E) and _Scn1a_fl/fl, _Emx1_-Cre (F) hippocampi, and 8-week-old _Scn1a_fl/fl (G) and _Scn1a_fl/fl, _Emx1_-Cre (H) neocortices, stained with anti-Nav1.1 antibody. Higher magnification images outlined in (a) are shown in (b). Scale bars: (a) 400 µm; (b) 40 µm. Arrowheads indicate the Nav1.1-immunoreactive bands extending from the stratum lacunosum-moleculare within the CA fields to the molecular layer of dentate gyrus, which virtually correspond to the hippocampal perforant path. DG, dentate gyrus; o, stratum oriens; p, stratum pyramidale; l, stratum lucidum; r, stratum radiatum; lm, stratum lacunosum-moleculare; m, dentate gyrus molecular layer; g, dentate gyrus granule cell layer. (I and J) Representative coronal sections of 8-week-old _Scn1a_fl/fl (I) and Scn1a_fl/fl, Emx1_-Cre (J) brains stained with anti-Nav1.1 antibody (n = 3 per genotype). Arrowheads and arrows indicate the Nav1.1-immunoreactive bands, which correspond to hippocampal perforant path and corpus callosum, respectively. Higher magnification images outlined in (a) are shown in (b). Note that reductions in Nav1.1 immunostaining intensities in the medial perforant pathway, corpus callosum and neocortical layers II/III, V and VI in _Scn1a_fl/fl, _Emx1_-Cre mice were observed in comparison with _Scn1a_fl/fl controls. CC, corpus callosum; DG, dentate gyrus; S1BF, primary somatosensory cortex, barrel field. Scale bars: 100 µm. Nuclear immunosignals are non-specific (15). Parasagittal images are oriented from the pial surface (top) to the callosal (bottom), and from rostral (left) to caudal (right). Coronal images are oriented from lateral (left) to medial (right). fl, floxed allele.
Figure 6.
Nav1.1 immunosignals in forebrain excitatory neurons are reduced in _Scn1a_fl/fl, _Emx1_-Cre mice. (A and B) Representative parasagittal sections of P21.5 _Scn1a_fl/fl (A) and _Scn1a_fl/fl, _Emx1_-Cre (B) hippocampi stained with anti-Nav1.1 antibody. Higher magnification images outlined in (a) are shown in (b). Scale bars: (a) 400 µm; (b) 40 µm. DG, dentate gyrus; o, stratum oriens; p, stratum pyramidale; l, stratum lucidum. (C and D) Representative parasagittal sections of P21.5 _Scn1a_fl/fl (C) and _Scn1a_fl/fl, _Emx1_-Cre (D) neocortices, stained with anti-Nav1.1 antibody. Higher magnification images outlined in (a) are shown in (b). Scale bars: (a) 400 µm; (b) 40 µm. Arrows indicate Nav1.1-immunoreactive neurites oriented toward the pial surface, which putatively correspond to AISs of neocortical PV cells (15,36) (see also Fig. 9O). Double arrowheads indicate Nav1.1-immunoreactive proximal neurites oriented toward the ventricular surface in neocortical layer V. (E–H) Representative parasagittal sections of 8-week-old _Scn1a_fl/fl (E) and _Scn1a_fl/fl, _Emx1_-Cre (F) hippocampi, and 8-week-old _Scn1a_fl/fl (G) and _Scn1a_fl/fl, _Emx1_-Cre (H) neocortices, stained with anti-Nav1.1 antibody. Higher magnification images outlined in (a) are shown in (b). Scale bars: (a) 400 µm; (b) 40 µm. Arrowheads indicate the Nav1.1-immunoreactive bands extending from the stratum lacunosum-moleculare within the CA fields to the molecular layer of dentate gyrus, which virtually correspond to the hippocampal perforant path. DG, dentate gyrus; o, stratum oriens; p, stratum pyramidale; l, stratum lucidum; r, stratum radiatum; lm, stratum lacunosum-moleculare; m, dentate gyrus molecular layer; g, dentate gyrus granule cell layer. (I and J) Representative coronal sections of 8-week-old _Scn1a_fl/fl (I) and Scn1a_fl/fl, Emx1_-Cre (J) brains stained with anti-Nav1.1 antibody (n = 3 per genotype). Arrowheads and arrows indicate the Nav1.1-immunoreactive bands, which correspond to hippocampal perforant path and corpus callosum, respectively. Higher magnification images outlined in (a) are shown in (b). Note that reductions in Nav1.1 immunostaining intensities in the medial perforant pathway, corpus callosum and neocortical layers II/III, V and VI in _Scn1a_fl/fl, _Emx1_-Cre mice were observed in comparison with _Scn1a_fl/fl controls. CC, corpus callosum; DG, dentate gyrus; S1BF, primary somatosensory cortex, barrel field. Scale bars: 100 µm. Nuclear immunosignals are non-specific (15). Parasagittal images are oriented from the pial surface (top) to the callosal (bottom), and from rostral (left) to caudal (right). Coronal images are oriented from lateral (left) to medial (right). fl, floxed allele.
Figure 7.
Additional _Emx1_-Cre-mediated Nav1.1 deletion in _Scn1a_fl/+, _VGAT_-Cre mice ameliorates seizure-related sudden death. Survival curves of P3 _Scn1a_fl/+, _Scn1a_fl/+, _Emx1_-Cre, _Scn1a_fl/+, _Emx1_-Cre,_VGAT_-Cre and _Scn1a_fl/+, _VGAT_-Cre littermates. Survival curve of _Scn1a_d/+ (reprinted from Fig. 1F) is included for comparison. Note that a significant decrease in premature lethality in _Scn1a_fl/+, _Emx1_-Cre,_VGAT_-Cre mice was observed in comparison with _Scn1a_fl/+, _VGAT_-Cre mice. +, wild-type allele; fl, floxed allele.
Figure 8.
Selective Nav1.1 deletion in mouse PV cells by _PV_-Cre causes spontaneous epileptic seizures and premature death. (A) Verification of _PV_-Cre-TG-dependent floxed Scn1a gene deletion in the brain by PCR analysis. (B and C) Verification of _PV_-Cre-TG-dependent Nav1.1 deletion by semi-quantitative western blot analysis on brain membrane proteins prepared from P21.5 _Scn1a_fl/fl,_PV_-Cre-TG and _Scn1a_fl/fl littermates, using anti-Nav1.1 antibody. Error bars represent SEM; two independent assays were performed. (D and E) Representative interictal (D) and ictal (E) ECoG recordings in P14–16 _Scn1a_fl/fl, _PV_-Cre-TG mice and _Scn1a_fl/fl control mice (n = 3, each group). (F) Survival curves of P3 _Scn1a_fl/fl, _Scn1a_fl/+, _Scn1a_fl/+, _PV_-Cre-TG and _Scn1a_fl/fl,_PV_-Cre-TG littermates. Note that all _Scn1a_fl/fl, _PV_-Cre-TG mice died before P25. +, wild-type allele; fl, floxed allele. ***P < 0.001.
Figure 9.
Nav1.1 immunosignals in PV inhibitory neurons are reduced at later developmental stages in _Scn1a_fl/fl, _PV_-TG-Cre mice. (A–N) Representative parasagittal sections of P12.5 _Scn1a_fl/fl (A) and _Scn1a_fl/fl, _PV-_Cre-TG (B) hippocampi, P12.5 _Scn1a_fl/fl (C) and _Scn1a_fl/fl, _PV-_Cre-TG (D) neocortices, P16.5 _Scn1a_fl/fl (E) and _Scn1a_fl/fl, _PV-_Cre-TG (F) hippocampi, P16.5 _Scn1a_fl/fl (G) and _Scn1a_fl/fl, _PV-_Cre-TG (H) neocortices, P21.5 _Scn1a_fl/fl (I) and _Scn1a_fl/fl, _PV-_Cre-TG (J) hippocampi and P21.5 _Scn1a_fl/fl (K and M) and _Scn1a_fl/fl, _PV-_Cre-TG (L and N) neocortices, stained with anti-Nav1.1 antibody. P21.5 was the oldest age used for analysis as _Scn1a_fl/fl, _PV_-TG-Cre mice show a high mortality rate beyond P15 and a life expectancy within P30. Higher magnification images outlined in (a) are shown in (b). Arrowheads indicate putative Nav1.1-immunoreactive AISs of neocortical PV cells. Double arrowheads indicate Nav1.1-immunoreactive proximal neurites oriented toward the ventricular surface. Scale bars: (a) 400 µm; (b) 40 µm. DG, dentate gyrus; o, stratum oriens; p, stratum pyramidale; l, stratum lucidum. (O and P) Immunofluorescence histochemistry of neocortices of P21.5 _Scn1a_fl/fl (O) and _Scn1a_fl/fl, _PV_-TG-Cre (P) mice stained with anti-Nav1.1 (a; red), PV (b; green) and ankyrin G (c; cyan) antibodies and counterstained with DAPI (d; blue). Merged images are shown (e). Arrowheads indicate AISs of neocortical PV cells with strong Nav1.1 immunofluorescence. An arrow indicates an AIS of a PV cell with reduced Nav1.1 immunofluorescence. Scale bars: 100 µm. (Q and R) Representative parasagittal sections of P12.5 _Scn1a_fl/fl (Q) and _Scn1a_fl/fl, _PV_-TG-Cre (R) cerebellum stained with anti-Nav1.1 antibody. Higher magnification images outlined in (a) are shown in (b) and (d). Higher magnification images outlined in (b) are shown in (c). Scale bars: (a) 400 µm; (b and c) 40 µm; (d) 80 µm. Arrowheads and arrows indicate Nav1.1-immunoreactive AISs of Purkinje cells and axons of basket cells, respectively. M, molecular cell layer; P, Purkinje cell layer; G, granule cell layer; CbN, cerebellar nuclei. Nuclear immunosignals are non-specific (15). Images are oriented from the pial surface (top) to the callosal (bottom), and from rostral (left) to caudal (right). fl, floxed allele.
Figure 9.
Nav1.1 immunosignals in PV inhibitory neurons are reduced at later developmental stages in _Scn1a_fl/fl, _PV_-TG-Cre mice. (A–N) Representative parasagittal sections of P12.5 _Scn1a_fl/fl (A) and _Scn1a_fl/fl, _PV-_Cre-TG (B) hippocampi, P12.5 _Scn1a_fl/fl (C) and _Scn1a_fl/fl, _PV-_Cre-TG (D) neocortices, P16.5 _Scn1a_fl/fl (E) and _Scn1a_fl/fl, _PV-_Cre-TG (F) hippocampi, P16.5 _Scn1a_fl/fl (G) and _Scn1a_fl/fl, _PV-_Cre-TG (H) neocortices, P21.5 _Scn1a_fl/fl (I) and _Scn1a_fl/fl, _PV-_Cre-TG (J) hippocampi and P21.5 _Scn1a_fl/fl (K and M) and _Scn1a_fl/fl, _PV-_Cre-TG (L and N) neocortices, stained with anti-Nav1.1 antibody. P21.5 was the oldest age used for analysis as _Scn1a_fl/fl, _PV_-TG-Cre mice show a high mortality rate beyond P15 and a life expectancy within P30. Higher magnification images outlined in (a) are shown in (b). Arrowheads indicate putative Nav1.1-immunoreactive AISs of neocortical PV cells. Double arrowheads indicate Nav1.1-immunoreactive proximal neurites oriented toward the ventricular surface. Scale bars: (a) 400 µm; (b) 40 µm. DG, dentate gyrus; o, stratum oriens; p, stratum pyramidale; l, stratum lucidum. (O and P) Immunofluorescence histochemistry of neocortices of P21.5 _Scn1a_fl/fl (O) and _Scn1a_fl/fl, _PV_-TG-Cre (P) mice stained with anti-Nav1.1 (a; red), PV (b; green) and ankyrin G (c; cyan) antibodies and counterstained with DAPI (d; blue). Merged images are shown (e). Arrowheads indicate AISs of neocortical PV cells with strong Nav1.1 immunofluorescence. An arrow indicates an AIS of a PV cell with reduced Nav1.1 immunofluorescence. Scale bars: 100 µm. (Q and R) Representative parasagittal sections of P12.5 _Scn1a_fl/fl (Q) and _Scn1a_fl/fl, _PV_-TG-Cre (R) cerebellum stained with anti-Nav1.1 antibody. Higher magnification images outlined in (a) are shown in (b) and (d). Higher magnification images outlined in (b) are shown in (c). Scale bars: (a) 400 µm; (b and c) 40 µm; (d) 80 µm. Arrowheads and arrows indicate Nav1.1-immunoreactive AISs of Purkinje cells and axons of basket cells, respectively. M, molecular cell layer; P, Purkinje cell layer; G, granule cell layer; CbN, cerebellar nuclei. Nuclear immunosignals are non-specific (15). Images are oriented from the pial surface (top) to the callosal (bottom), and from rostral (left) to caudal (right). fl, floxed allele.
Figure 10.
Minimal selective Nav1.1 deletion in PV cells by low-efficiency _PV_-Cre recombination is sufficient to cause spontaneous epileptic seizures and occasional premature death in mice. (A) Verification of _PV_-Cre-KI-dependent floxed Scn1a gene deletion in _Scn1a_fl/fl, _PV_-Cre-KI brain by PCR analysis. (B and C) Verification of _PV_-Cre-KI-dependent Nav1.1 deletion by semi-quantitative western blot analysis of brain membrane proteins prepared from P21.5 and 8-week-old _Scn1a_fl/fl, _PV_-Cre-KI and _Scn1a_fl/fl littermates, using anti-Nav1.1 antibody (B). Mean Nav1.1 expression levels in _Scn1a_fl/fl, _PV_-Cre-KI mice are represented as percentages relative to _Scn1a_fl/fl mice (100%) (C). Error bars represent SEM; two independent assays were performed. (D–F) Spontaneous epileptic seizures in _Scn1a_fl/fl, _PV_-Cre-KI mice. Representative normal interictal (D), ictal (E) and abnormal interictal (F) ECoGs recorded in _Scn1a_fl/fl, _PV_-Cre-KI mice are shown. ECoG recordings were performed with P25–30 _Scn1a_fl/fl, _PV_-Cre-KI and _Scn1a_fl/fl control mice. (G) Survival curves of P3 _Scn1a_fl/fl, _Scn1a_fl/+, _Scn1a_fl/+, _PV_-Cre-KI and _Scn1a_fl/fl, _PV_-Cre-KI littermates. A proportion of _Scn1a_fl/fl, _PV_-Cre-KI mice suffered sporadic death after P25. +, wild-type allele; fl, floxed allele.
Figure 11.
Nav1.1 immunosignals in PV inhibitory neurons are moderately reduced at later developmental stages in _Scn1a_fl/fl, _PV_-KI-Cre mice. (A–L) Representative parasagittal sections of P16.5 _Scn1a_fl/fl (A) and _Scn1a_fl/fl, _PV_-Cre-KI (B) hippocampi, P16.5 _Scn1a_fl/fl (C) and _Scn1a_fl/fl, _PV_-Cre-KI (D) neocortices, P21.5 _Scn1a_fl/fl (E) and _Scn1a_fl/fl, _PV_-Cre-KI (F) hippocampi, P21.5 _Scn1a_fl/fl (G) and _Scn1a_fl/fl, _PV_-Cre-KI (H) neocortices, 8-week-old _Scn1a_fl/fl (I) and _Scn1a_fl/fl, _PV_-Cre-KI (J) hippocampi and 8-week-old _Scn1a_fl/fl (K) and _Scn1a_fl/fl, _PV_-Cre-KI (L) neocortices, stained with anti-Nav1.1 antibody. Higher magnification images outlined in (a) are shown in (b). Scale bars: (a) 400 µm; (b) 40 µm. Arrowheads and double arrowheads indicate Nav1.1-immunoreactive AISs of neocortical PV cells and hippocampal perforant path afferents, respectively. DG, dentate gyrus; o, stratum oriens; p, stratum pyramidale; l, stratum lucidum; r, stratum radiatum; lm, stratum lacunosum-moleculare; m, dentate gyrus molecular layer; g, dentate gyrus granule cell layer. (M and N) Representative parasagittal sections of P21.5 _Scn1a_fl/fl (M) and _Scn1a_fl/fl, _PV_-Cre-KI (N) cerebellum, stained with anti-Nav1.1 antibody. Higher magnification images outlined in (a) are shown in (b) and (d). Higher magnification images outlined in (b) are shown in (c). Arrows indicate putative Nav1.1-immunoreactive axons of basket cells. Scale bars: (a) 400 µm; (b and c) 40 µm; (d) 80 µm. M, molecular cell layer; P, Purkinje cell layer; G, granule cell layer; CbN, cerebellar nuclei. Nuclear Nav1.1 immunostaining is non-specific (15). Images are oriented from the pial surface (top) to the callosal (bottom), and from rostral (left) to caudal (right). fl, floxed allele.
Figure 11.
Nav1.1 immunosignals in PV inhibitory neurons are moderately reduced at later developmental stages in _Scn1a_fl/fl, _PV_-KI-Cre mice. (A–L) Representative parasagittal sections of P16.5 _Scn1a_fl/fl (A) and _Scn1a_fl/fl, _PV_-Cre-KI (B) hippocampi, P16.5 _Scn1a_fl/fl (C) and _Scn1a_fl/fl, _PV_-Cre-KI (D) neocortices, P21.5 _Scn1a_fl/fl (E) and _Scn1a_fl/fl, _PV_-Cre-KI (F) hippocampi, P21.5 _Scn1a_fl/fl (G) and _Scn1a_fl/fl, _PV_-Cre-KI (H) neocortices, 8-week-old _Scn1a_fl/fl (I) and _Scn1a_fl/fl, _PV_-Cre-KI (J) hippocampi and 8-week-old _Scn1a_fl/fl (K) and _Scn1a_fl/fl, _PV_-Cre-KI (L) neocortices, stained with anti-Nav1.1 antibody. Higher magnification images outlined in (a) are shown in (b). Scale bars: (a) 400 µm; (b) 40 µm. Arrowheads and double arrowheads indicate Nav1.1-immunoreactive AISs of neocortical PV cells and hippocampal perforant path afferents, respectively. DG, dentate gyrus; o, stratum oriens; p, stratum pyramidale; l, stratum lucidum; r, stratum radiatum; lm, stratum lacunosum-moleculare; m, dentate gyrus molecular layer; g, dentate gyrus granule cell layer. (M and N) Representative parasagittal sections of P21.5 _Scn1a_fl/fl (M) and _Scn1a_fl/fl, _PV_-Cre-KI (N) cerebellum, stained with anti-Nav1.1 antibody. Higher magnification images outlined in (a) are shown in (b) and (d). Higher magnification images outlined in (b) are shown in (c). Arrows indicate putative Nav1.1-immunoreactive axons of basket cells. Scale bars: (a) 400 µm; (b and c) 40 µm; (d) 80 µm. M, molecular cell layer; P, Purkinje cell layer; G, granule cell layer; CbN, cerebellar nuclei. Nuclear Nav1.1 immunostaining is non-specific (15). Images are oriented from the pial surface (top) to the callosal (bottom), and from rostral (left) to caudal (right). fl, floxed allele.
Similar articles
- Antisense oligonucleotides restore excitability, GABA signalling and sodium current density in a Dravet syndrome model.
Yuan Y, Lopez-Santiago L, Denomme N, Chen C, O'Malley HA, Hodges SL, Ji S, Han Z, Christiansen A, Isom LL. Yuan Y, et al. Brain. 2024 Apr 4;147(4):1231-1246. doi: 10.1093/brain/awad349. Brain. 2024. PMID: 37812817 Free PMC article. - Targeted Augmentation of Nuclear Gene Output (TANGO) of Scn1a rescues parvalbumin interneuron excitability and reduces seizures in a mouse model of Dravet Syndrome.
Wengert ER, Wagley PK, Strohm SM, Reza N, Wenker IC, Gaykema RP, Christiansen A, Liau G, Patel MK. Wengert ER, et al. Brain Res. 2022 Jan 15;1775:147743. doi: 10.1016/j.brainres.2021.147743. Epub 2021 Nov 26. Brain Res. 2022. PMID: 34843701 - Genetic background modulates impaired excitability of inhibitory neurons in a mouse model of Dravet syndrome.
Rubinstein M, Westenbroek RE, Yu FH, Jones CJ, Scheuer T, Catterall WA. Rubinstein M, et al. Neurobiol Dis. 2015 Jan;73:106-17. doi: 10.1016/j.nbd.2014.09.017. Epub 2014 Oct 2. Neurobiol Dis. 2015. PMID: 25281316 Free PMC article. - From genotype to phenotype in Dravet disease.
Gataullina S, Dulac O. Gataullina S, et al. Seizure. 2017 Jan;44:58-64. doi: 10.1016/j.seizure.2016.10.014. Epub 2016 Oct 21. Seizure. 2017. PMID: 27817982 Review. - Molecular and cellular basis: insights from experimental models of Dravet syndrome.
Yamakawa K. Yamakawa K. Epilepsia. 2011 Apr;52 Suppl 2:70-1. doi: 10.1111/j.1528-1167.2011.03006.x. Epilepsia. 2011. PMID: 21463284 Review.
Cited by
- Epigenetic insights into GABAergic development in Dravet Syndrome iPSC and therapeutic implications.
Schuster J, Lu X, Dang Y, Klar J, Wenz A, Dahl N, Chen X. Schuster J, et al. Elife. 2024 Aug 27;12:RP92599. doi: 10.7554/eLife.92599. Elife. 2024. PMID: 39190448 Free PMC article. - Functional Characteristics of the Nav1.1 p.Arg1596Cys Mutation Associated with Varying Severity of Epilepsy Phenotypes.
Witkowski G, Szulczyk B, Nurowska E, Jurek M, Pasierski M, Lipiec A, Charzewska A, Dawidziuk M, Milewski M, Owsiak S, Rola R, Sienkiewicz Jarosz H, Hoffman-Zacharska D. Witkowski G, et al. Int J Mol Sci. 2024 Feb 1;25(3):1745. doi: 10.3390/ijms25031745. Int J Mol Sci. 2024. PMID: 38339022 Free PMC article. - Preferential expression of SCN1A in GABAergic neurons improves survival and epileptic phenotype in a mouse model of Dravet syndrome.
Ricobaraza A, Bunuales M, Gonzalez-Aparicio M, Fadila S, Rubinstein M, Vides-Urrestarazu I, Banderas J, Sola-Sevilla N, Sanchez-Carpintero R, Lanciego JL, Roda E, Honrubia A, Arnaiz P, Hernandez-Alcoceba R. Ricobaraza A, et al. J Mol Med (Berl). 2023 Dec;101(12):1587-1601. doi: 10.1007/s00109-023-02383-8. Epub 2023 Oct 11. J Mol Med (Berl). 2023. PMID: 37819378 Free PMC article. - Antisense oligonucleotides restore excitability, GABA signalling and sodium current density in a Dravet syndrome model.
Yuan Y, Lopez-Santiago L, Denomme N, Chen C, O'Malley HA, Hodges SL, Ji S, Han Z, Christiansen A, Isom LL. Yuan Y, et al. Brain. 2024 Apr 4;147(4):1231-1246. doi: 10.1093/brain/awad349. Brain. 2024. PMID: 37812817 Free PMC article. - Development of two mouse strains conditionally expressing bright luciferases with distinct emission spectra as new tools for in vivo imaging.
Nakashiba T, Ogoh K, Iwano S, Sugiyama T, Mizuno-Iijima S, Nakashima K, Mizuno S, Sugiyama F, Yoshiki A, Miyawaki A, Abe K. Nakashiba T, et al. Lab Anim (NY). 2023 Oct;52(10):247-257. doi: 10.1038/s41684-023-01238-6. Epub 2023 Sep 7. Lab Anim (NY). 2023. PMID: 37679611 Free PMC article.
References
- Escayg A., MacDonald B.T., Meisler M.H., Baulac S., Huberfeld G., An-Gourfinkel I., Brice A., LeGuern E., Moulard B., Chaigne D., et al. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat. Genet. 2000;24:343–345. doi:10.1038/74159. - DOI - PubMed
- Claes L., Del-Favero J., Ceulemans B., Lagae L., Van Broeckhoven C., De Jonghe P. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am. J. Hum. Genet. 2001;68:1327–1332. doi:10.1086/320609. - DOI - PMC - PubMed
- Sugawara T., Mazaki-Miyazaki E., Ito M., Nagafuji H., Fukuma G., Mitsudome A., Wada K., Kaneko S., Hirose S., Yamakawa K. Nav1.1 mutations cause febrile seizures associated with afebrile partial seizures. Neurology. 2001;57:703–705. doi:10.1212/WNL.57.4.703. - DOI - PubMed
- Sugawara T., Mazaki-Miyazaki E., Fukushima K., Shimomura J., Fujiwara T., Hamano S., Inoue Y., Yamakawa K. Frequent mutations of SCN1A in severe myoclonic epilepsy in infancy. Neurology. 2002;58:1122–1124. doi:10.1212/WNL.58.7.1122. - DOI - PubMed
- Fujiwara T., Sugawara T., Mazaki-Miyazaki E., Takahashi Y., Fukushima K., Watanabe M., Hara K., Morikawa T., Yagi K., Yamakawa K., et al. Mutations of sodium channel α subunit type 1 (SCN1A) in intractable childhood epilepsies with frequent generalized tonic-clonic seizures. Brain. 2003;126:531–546. doi:10.1093/brain/awg053. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases