Charting a dynamic DNA methylation landscape of the human genome - PubMed (original) (raw)
. 2013 Aug 22;500(7463):477-81.
doi: 10.1038/nature12433. Epub 2013 Aug 7.
Hongcang Gu, Fabian Müller, Julie Donaghey, Linus T-Y Tsai, Oliver Kohlbacher, Philip L De Jager, Evan D Rosen, David A Bennett, Bradley E Bernstein, Andreas Gnirke, Alexander Meissner
Affiliations
- PMID: 23925113
- PMCID: PMC3821869
- DOI: 10.1038/nature12433
Charting a dynamic DNA methylation landscape of the human genome
Michael J Ziller et al. Nature. 2013.
Abstract
DNA methylation is a defining feature of mammalian cellular identity and is essential for normal development. Most cell types, except germ cells and pre-implantation embryos, display relatively stable DNA methylation patterns, with 70-80% of all CpGs being methylated. Despite recent advances, we still have a limited understanding of when, where and how many CpGs participate in genomic regulation. Here we report the in-depth analysis of 42 whole-genome bisulphite sequencing data sets across 30 diverse human cell and tissue types. We observe dynamic regulation for only 21.8% of autosomal CpGs within a normal developmental context, most of which are distal to transcription start sites. These dynamic CpGs co-localize with gene regulatory elements, particularly enhancers and transcription-factor-binding sites, which allow identification of key lineage-specific regulators. In addition, differentially methylated regions (DMRs) often contain single nucleotide polymorphisms associated with cell-type-related diseases as determined by genome-wide association studies. The results also highlight the general inefficiency of whole-genome bisulphite sequencing, as 70-80% of the sequencing reads across these data sets provided little or no relevant information about CpG methylation. To demonstrate further the utility of our DMR set, we use it to classify unknown samples and identify representative signature regions that recapitulate major DNA methylation dynamics. In summary, although in theory every CpG can change its methylation state, our results suggest that only a fraction does so as part of coordinated regulatory programs. Therefore, our selected DMRs can serve as a starting point to guide new, more effective reduced representation approaches to capture the most informative fraction of CpGs, as well as further pinpoint putative regulatory elements.
Figures
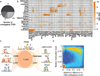
Figure 1. Identification and characteristics of differentially methylated regions (DMRs) in the human genome
a. Principal component analysis based on CpG methylation levels for 1kb tiles across 30 diverse human cell and tissue samples. Coloring indicates classification of samples into subgroups and group wise mean DNAme. Detailed sample annotations are listed in Supplementary Table 1. Gray area indicates Alzheimer’s disease (AD) samples.. b. Density scatterplot of CpG wise DNAme level differences (x-axis, p≤0.01) and CpG median methylation (y-axis) across the 24 developmental samples (excluding cancer and long-term culture). Coloring indicates CpG density from low (blue) to high (red). The red box highlights dynamic CpGs (≥0.3). c. Cumulative distribution of DMR specificity. High hypo/hypermethylation specificity indicates that particular region is methylated/unmethylated in most tissues and deviates from this default state in only one or few cases. d. Top: Composite plot of mean DNAme differences across various genomic features. Black line indicates the median of the average DNAme difference across each feature. Grey areas mark 25th and 75th percentile. Bottom: Distribution of mean DNAme difference for each genomic feature. Black bar indicates 25th and 75th percentile while white dot marks the median. For CGI islands, a smaller, experimentally determined set (eCGI; n=25,490) is shown as well. Promoters are broken down into high CpG content (HCP, n=24,899), intermediate CpG content (ICP, n=10,920) and low CpG content (LCP, n=7,946) regions (n=43,765 total). e. Methylation level variation across the OCT4 locus (chr6:31,119,000–31,162,000) (top). Blue boxes indicate DMRs significant at p≤0.01 and exhibit a minimum difference ≥0.3 across the 24 developmental samples. For reference, ENCODE TFBS cluster track, DNAse I hypersensitive sites, CpG islands and RefSeq genes are shown. f. Distribution of DMRs across various genomic features. Each region is assigned only to one of these genomic feature according the ranking promoter, CGI, CGI shore, exon, intron, putative enhancers, DNAse I hypersensitive site or other.
Figure 2. Dynamic CpG methylation regions frequently co-localize with transcription factor binding sites (TFBS)
a. Overlap of DMRs with ENCODE TFBS. b. Enrichment of the top four TFBSs significantly overrepresented (p<0.01, empirical test) in DMRs specific to the cell type indicated (specificity >0.15). Color code quantifies median enrichment odds ratio compared to size matched random control regions. c. Overlap of PAX5 motifs (±100bp top) unmethylated in CD34 cells or fetal brain across the entire human genome. Regions specifically unmethylated in CD34 or fetal brain were subjected to motif analysis and top differentially co-occurring motifs are highlighted on the left for CD34 and on the right for fetal brain. d. Density scatterplot of maximum DNAme difference across 24 developmental samples for TFBS cluster track (n=2.7 million) and median methylation level across all samples. Color code indicates density of TFBS from low (blue) to high (red)..
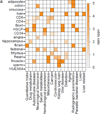
Figure 3. DMRs exhibit elevated SNP frequency and show non-random GWAS SNP enrichment
a. Odds ratio of significantly overrepresented (p<0.05, empirical test, see Supplementary Information) GWAS SNPs grouped into 16 categories in regions specifically hypomethylated within the sample indicated on the left. Asterisk indicates p-value <0.1.
Figure 4. Effective classification and sample deconvolution using only the DMR set
a. Overlap of dynamic CpGs (p≤0.01 Δ≥0.3) in normal samples and between colon cancer and matching control CpG numbers (in million). b. Distribution of autosomal CpGs across three conditions. Class name indicates sample group where a CpG was observed dynamic (developmental (n=24), cell culture (n=3), cancer (n=2)) or remained unchanged over the entire sample set (n=30). c. Repeat content distribution of DMRs (sets as in b). d. Hierarchical clustering using pearson correlation coefficient (PCC) of the DMR values across the entire sample set (n=30). e. Distance of the fetal brain sample to different sets of signature regions defined for sample classes or individual samples, but excluding regions identified by means of the fetal brain sample. f. Contribution of individual sample signature region sets to an in silico generated hybrid sample (HUES64 and hippocampus).
Similar articles
- Mapping of Variable DNA Methylation Across Multiple Cell Types Defines a Dynamic Regulatory Landscape of the Human Genome.
Gu J, Stevens M, Xing X, Li D, Zhang B, Payton JE, Oltz EM, Jarvis JN, Jiang K, Cicero T, Costello JF, Wang T. Gu J, et al. G3 (Bethesda). 2016 Apr 7;6(4):973-86. doi: 10.1534/g3.115.025437. G3 (Bethesda). 2016. PMID: 26888867 Free PMC article. - Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling.
Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, Van Djik S, Muhlhausler B, Stirzaker C, Clark SJ. Pidsley R, et al. Genome Biol. 2016 Oct 7;17(1):208. doi: 10.1186/s13059-016-1066-1. Genome Biol. 2016. PMID: 27717381 Free PMC article. - Elucidating the landscape of aberrant DNA methylation in hepatocellular carcinoma.
Song MA, Tiirikainen M, Kwee S, Okimoto G, Yu H, Wong LL. Song MA, et al. PLoS One. 2013;8(2):e55761. doi: 10.1371/journal.pone.0055761. Epub 2013 Feb 20. PLoS One. 2013. PMID: 23437062 Free PMC article. - Genome-scale DNA methylation analysis.
Fouse SD, Nagarajan RO, Costello JF. Fouse SD, et al. Epigenomics. 2010 Feb;2(1):105-17. doi: 10.2217/epi.09.35. Epigenomics. 2010. PMID: 20657796 Free PMC article. Review. - Reshaping the Landscape of the Genome: Toolkits for Precise DNA Methylation Manipulation and Beyond.
Zhu C, Hao Z, Liu D. Zhu C, et al. JACS Au. 2023 Dec 21;4(1):40-57. doi: 10.1021/jacsau.3c00671. eCollection 2024 Jan 22. JACS Au. 2023. PMID: 38274248 Free PMC article. Review.
Cited by
- A comprehensive lifestyle index and its associations with DNA methylation and type 2 diabetes among Ghanaian adults: the rodam study.
Abidha CA, Meeks KAC, Chilunga FP, Venema A, Schindlmayr R, Hayfron-Benjamin C, Klipstein-Grobusch K, Mockenhaupt FP, Agyemang C, Henneman P, Danquah I. Abidha CA, et al. Clin Epigenetics. 2024 Oct 16;16(1):143. doi: 10.1186/s13148-024-01758-z. Clin Epigenetics. 2024. PMID: 39415250 Free PMC article. - The role of DNA methylation in chondrogenesis of human iPSCs as a stable marker of cartilage quality.
Hajmousa G, de Almeida RC, Bloks N, Ruiz AR, Bouma M, Slieker R, Kuipers TB, Nelissen RGHH, Ito K, Freund C, Ramos YFM, Meulenbelt I. Hajmousa G, et al. Clin Epigenetics. 2024 Oct 15;16(1):141. doi: 10.1186/s13148-024-01759-y. Clin Epigenetics. 2024. PMID: 39407288 Free PMC article. - Improved detection of methylation in ancient DNA.
Sawyer S, Gelabert P, Yakir B, Llanos-Lizcano A, Sperduti A, Bondioli L, Cheronet O, Neugebauer-Maresch C, Teschler-Nicola M, Novak M, Pap I, Szikossy I, Hajdu T, Moiseyev V, Gromov A, Zariņa G, Meshorer E, Carmel L, Pinhasi R. Sawyer S, et al. Genome Biol. 2024 Oct 10;25(1):261. doi: 10.1186/s13059-024-03405-5. Genome Biol. 2024. PMID: 39390557 Free PMC article. - Methylation patterns at the adjacent CpG sites within enhancers are a part of cell identity.
Taryma-Leśniak O, Bińkowski J, Przybylowicz PK, Sokolowska KE, Borowski K, Wojdacz TK. Taryma-Leśniak O, et al. Epigenetics Chromatin. 2024 Oct 10;17(1):30. doi: 10.1186/s13072-024-00555-5. Epigenetics Chromatin. 2024. PMID: 39385277 Free PMC article. - Implementation of the Methyl-Seq platform to identify tissue- and sex-specific DNA methylation differences in the rat epigenome.
Cox OH, Seifuddin F, Guo J, Pirooznia M, Boersma GJ, Wang J, Tamashiro KLK, Lee RS. Cox OH, et al. Epigenetics. 2024 Dec;19(1):2393945. doi: 10.1080/15592294.2024.2393945. Epub 2024 Sep 22. Epigenetics. 2024. PMID: 39306700 Free PMC article.
References
- Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. - PubMed
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RF1 AG036042/AG/NIA NIH HHS/United States
- P01GM099117/GM/NIGMS NIH HHS/United States
- R01AG15819/AG/NIA NIH HHS/United States
- ES017690/ES/NIEHS NIH HHS/United States
- P30AG10161/AG/NIA NIH HHS/United States
- U01 ES017155/ES/NIEHS NIH HHS/United States
- R01 AG015819/AG/NIA NIH HHS/United States
- R01 DK087092/DK/NIDDK NIH HHS/United States
- R01AG36042/AG/NIA NIH HHS/United States
- P01 GM099117/GM/NIGMS NIH HHS/United States
- U01ES017155/ES/NIEHS NIH HHS/United States
- R01 AG017917/AG/NIA NIH HHS/United States
- R01AG17917/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources